FOD-Net: A Deep Learning Method for Fiber Orientation Distribution Angular Super Resolution
Highlights
- A deep-learning-based framework for fibre orientation distribution (FOD) angular super-resolution in diffusion MRI (dMRI) data.
- The work improves the FOD angular resolution of FOD images acquired with single-shell low-angular-resolution dMRI data to approximate the quality of multi-shell high-angular-resolution dMRI FODs.
- Achieves super-resolution without requiring high b-values.
- Authors demonstrate the benefits of their super-resolved FODs in tractography and connectomics.
Introduction
The fiber orientation distribution is optimally estimated from high angular resolution diffusion imaging (HARDI) data. However, acquiring HARDI data is time consuming, and not frequent in clinical practice. Thus, the accuracy of the tractography products computed using clinical data is limited.
Authors propose a FOD network (FOD-Net), a deep-learning-based FOD angular super resolution method that directly enhances FOD data from single-shell low angular resolution imaging data to obtain the super-resolved FOD data equivalent to those derived from high-quality multi-shell HARDI acquisitions.
Methods
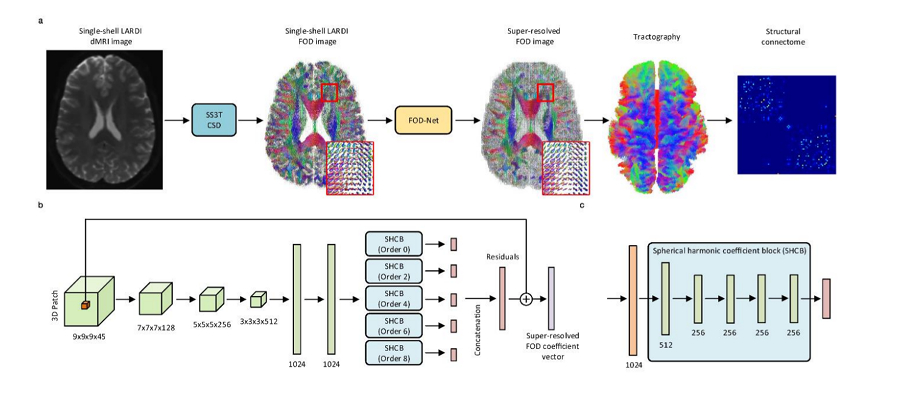
Authors use of a 3D convolutional neural network (CNN) as their model.
The FOD-Net architecture consists of:
- 5 convolutional layers
- 2 fully connected layers
- 5 five spherical harmonic coefficient blocks (SHCB) (one per SH expansion order).
The input to the network are the SH expansion coefficients, which can be set to a fixed length regardless of the number of gradient encoding directions. The network outputs the super-resolved version of the central voxel of the input patch.
The coefficients are normalized to avoid high order SH terms to have values close to zero. The data is split into fixed size patches to feed it to the network.
Data
- 110 subjects from the Human Connectome Project (HCP): the original multi-shell acquisition data was used as to generate the multi-shell multi-tissue (MSMT) ground truth FODs, and the single-shell three-tissue (SS3T) method applied on a subsampled version of the data was used as the signal to compute the FODs and feed the network.
- Local clinical 3 T scanner for generalization.
Results
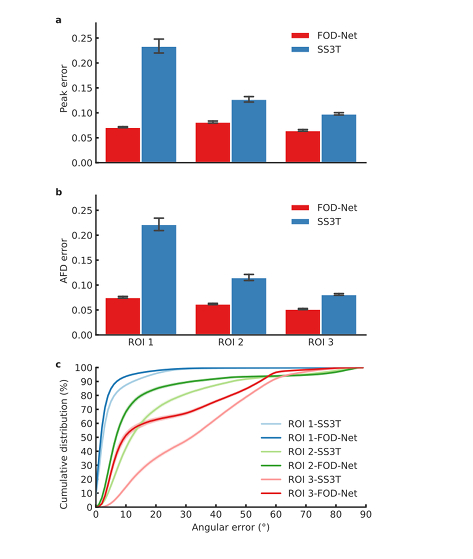
- FOD-Net yields larger improvements in crossing-fibre regions than coherent single fiber regions, which indicates that ambiguities generated by a high number of crossing-fibre voxels were successfully resolved by FOD-Net.
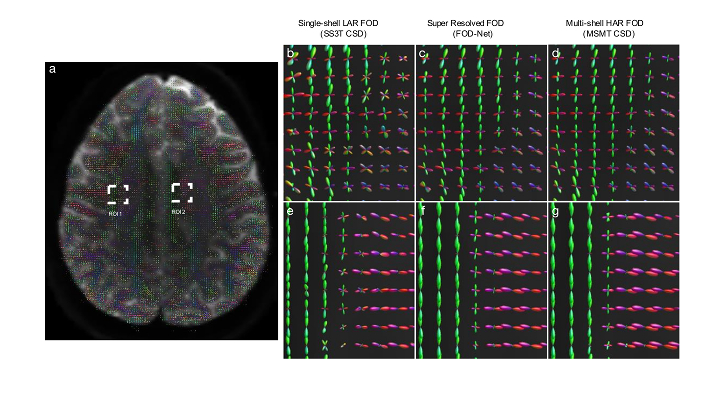
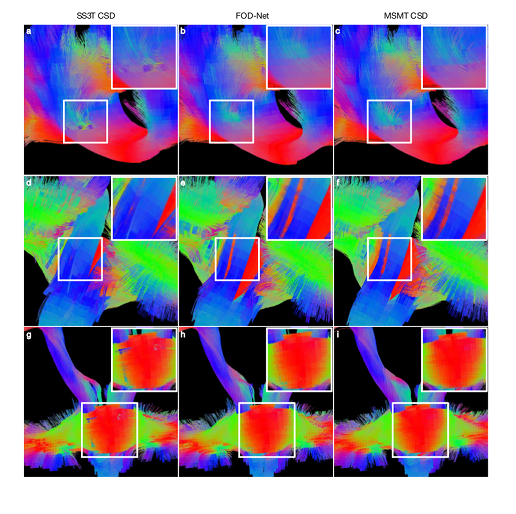
- FOD-Net removes spurious fibers and recovers missing fiber tracts in anatomical regions of interest.
- More reliable connectomes are generated by the enhanced fiber orientation map.
- FOD-Net has good generalization capability.
- The method is reproducible at test-retest.
Conclusions
FOD-Net produces FODs of equivalent quality to those produced from more advanced multi-shell HARDI data, but requires only a fraction of the dMRI data.
The use of SH coefficient patches provides the network some spatial context information, and might be thought as compensating the lack of angular contrast.
Comments
-
Being superior to SS3T-computed FODs is expected if the network learns to map to MSMT-computed FODs.
-
While not the first work using low angular resolution data to predict super-resolution FODs (or DTI) (see e.g. here, here, or here, or here, or other works authors cite whose purpose is the same using different input data or exactly the same SH coefficients), the work is interesting and provides extensive evidence about the feasibility of the approach for various tractography tasks.
Peak error computed by the absolute difference between the amplitude of a FOD estimate at the maximum peak and its corresponding ‘ground-truth’.
- Not sure how that explains that the error is lower for fiber crossing, which seems counter-intuitive, unless a given error is distributed/averaged across the multiple peaks, which is not implied by the statement.
- FOD-Net removes spurious fibers and recovers missing fiber tracts in anatomical regions of interest.
-
This is unclear; the network does not remove them per se, since the learned FODs resolve fiber crossings better, this is built-in or a by-product.
-
It would have been informative to measure the quality (e.g. OR, OL, etc.) of the produced tractograms.
-
It is unclear why authors used TractSeg for the tractography analysis but then use iFOD2 for the connectome analysis. It is also unclear if they applied SIFT2 to the tractograms generated with FODNet, which may weaken the previous statement about removing spurious fibers or might hinder to asses FODNet’s effect on the connectome since SIFT2 weighs the streamlines based on the information provided FOD: if FODs are better, then there would be less need of using SIFT2.
-
The method takes two weeks to converge (on 2 Tesla V100 GPUs).