Superfast Diffusion Tensor Imaging and Fiber Tractography Using Deep Learning
Highlights
Authors present a deep learning method to obtain diffusion quantitative maps and fiber tractography using a reduced set of gradient encoding directions.
Introduction
Diffusion measures usually require a number of diffusion encoding gradient directions of around 30 to provide diagnostic quality. Acquiring more diffusion gradient encoding directions requires longer scan time. Authors propose to use a deep convolutional neural network to predict reliably diffusion derived maps and tractography from a reduced set of gradient encoding directions.
Methods
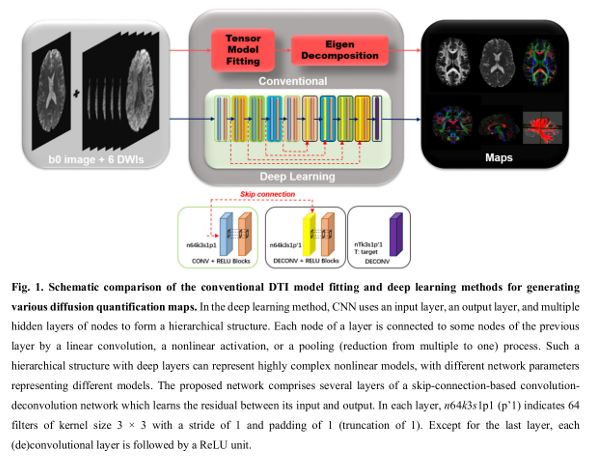
Authors propose a deep network to model the nonlinear relationship between the acquired diffusion data and the desired diffusion derived maps.
The network is a conventional ResNet. The input diffusion data is divided into two-dimensional overlapping patches to feed the network.
A Diffusion Tensor Imaging-like acquisition (6 diffusion volumes and an additional \(b0\) acquisition) are used as the input to the network.
The predicted diffusion maps are the median diffusivity (MD) and fractional anisotropy (FA).
The proposed model was also used to predict the FA maps on non-healthy individuals being trained only on healthy individuals.
Data
50 healthy individual data from the Human Connectome Project (HCP) was used for training and 10 subjects were used for testing. The HCP data used corresponds to the \(b = 1000 s/mm^2\) shell and 90 diffusion encoding gradient directions.
For testing the accuracy of the network on non-healthy individuals a separate dataset containing stroke patients was used.
Results
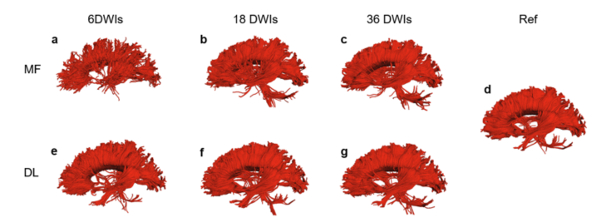
For the HCP tests, their method was compared to the MD, FA and tractography computed using “conventional tensor model fitting methods”. The reference contained information of all 90 diffusion volumes and the comparison was done on data using 8, 16, and 36 diffusion volumes.
The authors claim that their method achieves a lower error than the conventional tensor model fitting methods.
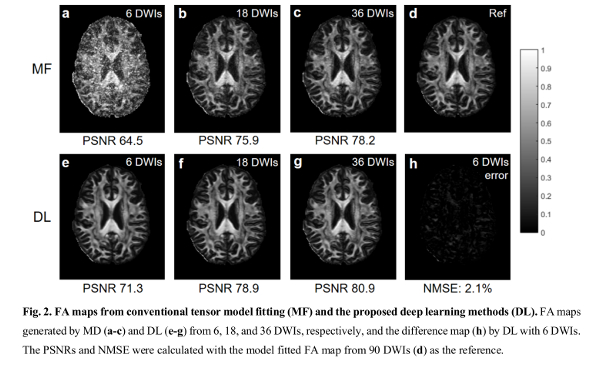
In an additional experiment, authors added Rician noise to the data to compare the robustness of their method to data imperfections.
For the non-healthy individuals’ dataset, the authors claim that the network is able to predict accurately the FA maps of non-healthy individuals with the model being trained only on healthy subject data.
Conclusions
Authors conclude that their deep learning model is able to accurately predict the MD and FA maps and tractography using a reduced set of diffusion volumes compared to “conventional tensor model fitting methods”.
Comments
- It is unclear the benefit of their approach concerning the FA and MD maps since these are computed using only 6 diffusion DTI-like data. It is unclear whether by FA for the 90 HCP directions they meant the GFA computed using spherical harmonic decomposition measures.
- As authors note, since they are using a DTI approach, the reference tractography is unable to resolve fiber crossings. Thus, using DL with more diffusion volumes is unlikely to bring any benefit or to learn crossing that are not in their ground truth.
- It is unclear the innovation of this work with respect of other DL methods that have been proposed for the same task. The comparison to such methods is missing.