Improving Explainability of Disentangled Representations using Multipath-Attribution Mappings
Highlights
- Leverage existing SOTA disentangled representation method (\(\beta\)-TCVAE) to learn interpretable representation for downstream tasks;
- Introduce a new multi-path attribution mapping to explain what guided a downstream networks’ prediction (the attribution w.r.t. the input), but also why it had an impact (the attribution w.r.t. the interpretable representation);
- The method helps to extract causal relations, as well as improves robustness to distribution shifts by detecting “shortcuts” learned by models.
Introduction
The authors argue that current approaches in interpretable deep learning are too prone to interpret correlation as causation. Ultimately, wrongly identifying the underlying causal relations also affects generalization capabilities, since it makes models more vulnerable to domain shifts, by correlating non-causal factors of variation to the prediction target.
Thus, the authors’ motivation is to provide a better framework to interpret correlations in the data, while leaving the task to discriminate between correlation and causation to domain experts.
Methods
The overall method is summarized in the figure below, and explained in more details in the following subsections:
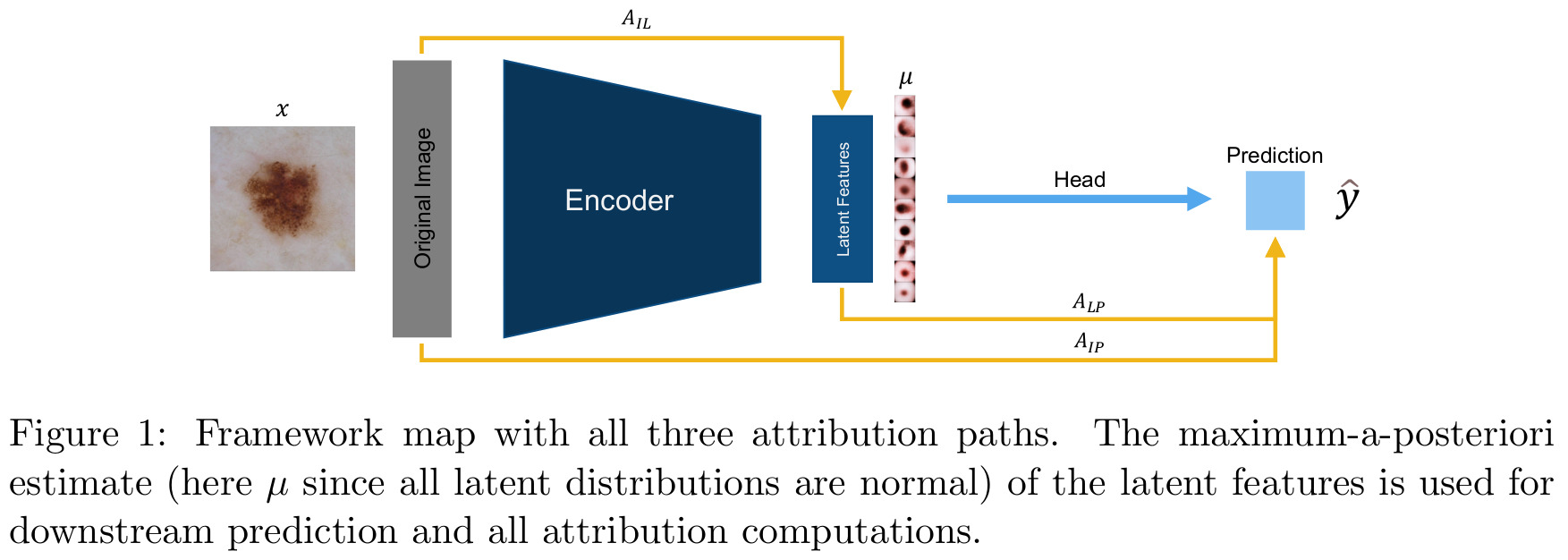
Autoencoder
To learn human-interpretable factors of variation to be used later in the attribution mechanism, the authors use the \(\beta\)-TCVAE1. Very briefly, \(\beta\)-TCVAE is a method that decomposes the KL divergence in VAEs into 3 terms. Amongst the 3 terms, the most notable is the total correlation of the aggregate posterior:
\[KL(q(z)~\|~\prod_{j=1}^{d} q(z_{j})), \quad \text{with}~d~\text{the number of latent dimensions}\]which can be penalized more heavily to promote a disentangled latent space, without any additional supervision.
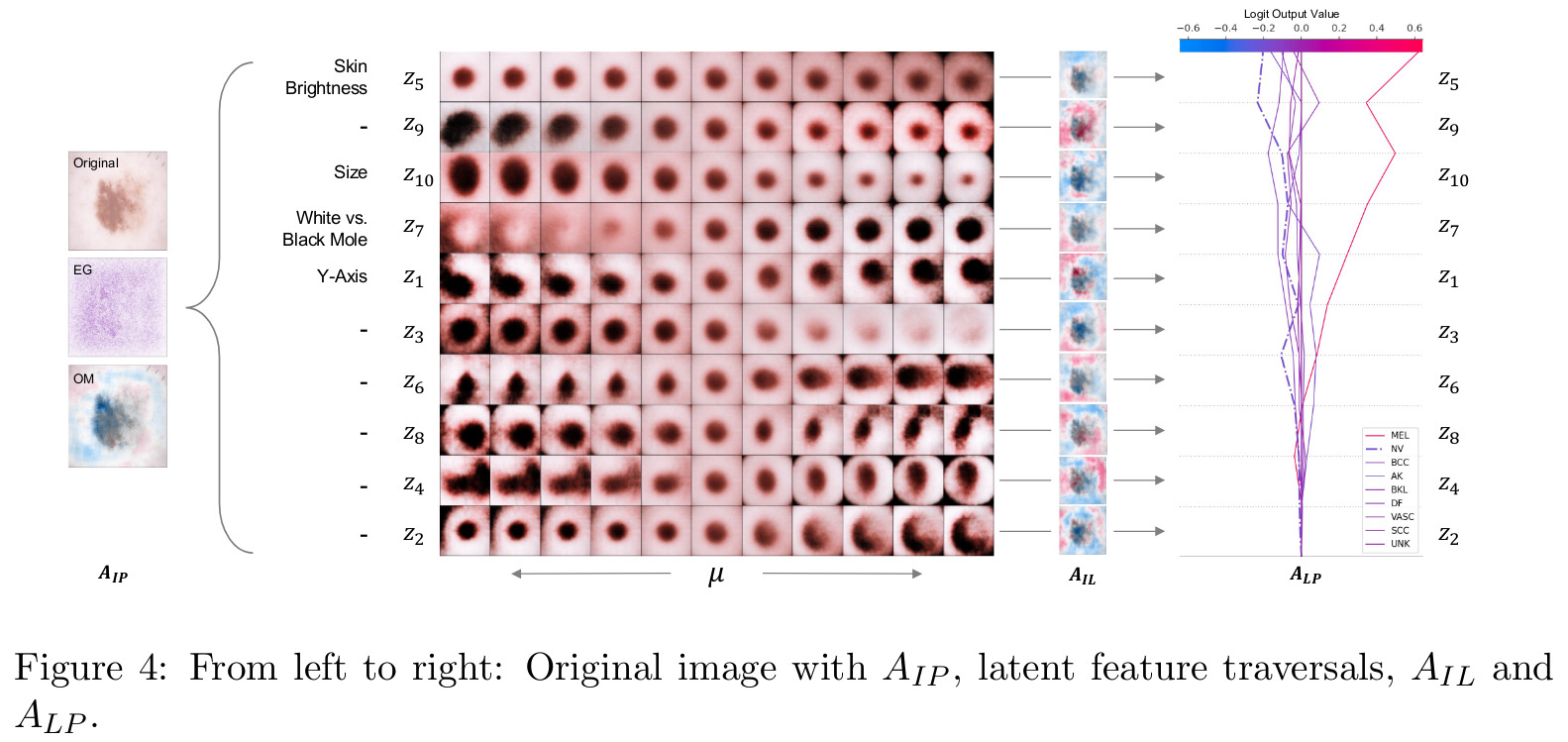
A posteriori, the authors visually inspected traversals of individual latent dimensions to manually associate interpretable generative factors to as many of the latent dimensions as possible.
Attribution methods
The authors mention multiple saliency methods for measuring the contribution of each \(x_i\) dimension of the input to a model’s predicting. Here, the authors use so called “perturbation methods”, which perturb the input and measure the effect on the prediction, namely the expected gradients (EG)2 and occlusion maps (OM)3.
The novel contribution of the paper is to introduce new attribution pathways, besides the standard pixel-wise image-into-prediction (\(A_{IP}\)) path:
- Latent-into-prediction \(A_{LP}\): Reveals what interpretable features where important for the prediction;
- Image-into-latent \(A_{IL}\): Sanity check of the method, to ensure that the captured factor of variation overlaps with its anticipated feature in the image.
Data
The authors evaluate their method on:
- one synthetic dataset based on MNIST (for a proof of concept);
- OCT Retina Scans dataset4, containing healthy and ill patients with three diseases;
- Skin Lesion Images5, containing dermoscopic images with nine different diagnostic categories.
Results
The authors present a huge amount of supplementary results, but here we’ll focus on the synthetic dataset and one of the medical imaging datasets.
Their results on the synthetic dataset help understand the various attribution paths.

On the skin lesion dataset, the authors use their method to interpret that the skin brightness and size were most responsible for leading the model to mistakenly predict a mole as a melanoma.
This is only mentioned in the supplementary materials, but their method’s interpretability/performance trade-off is substantial. In the interpretable settings, the best accuracies they achieved were of 95% for the synthetic dataset and 55-60% for the OCT dataset.
Using the same architecture, but allowing to finetune the encoder along with the MLP classification head (abandoning the latent dimensions’ interpretability in the process), they were able to reach accuracies of 96% for the synthetic dataset and 83% for the OCT dataset.
References
- Code is available on GitHub: https://github.com/IML-DKFZ/m-pax_lib
- A short video presentation is available on MIDL’s website: https://2022.midl.io/papers/b_l_12
-
Review of \(\beta\)-TCVAE: https://vitalab.github.io/article/2020/08/14/IsolatingSourcesOfDisentanglementInVAEs.html ↩
-
Erion et al., “Improving performance of deep learning models with axiomatic attribution priors and expected gradients”, Nature Machine Intelligence, 2021: https://arxiv.org/abs/1906.10670 ↩
-
Zeiler and Fergus, “Visualizing and Understanding Convolutional Networks”, ECCV, 2014: https://arxiv.org/abs/1311.2901 ↩
-
Kermany et al. “Identifying Medical Diagnoses and Treatable Diseases by Image-Based Deep Learning”, Cell, 2018: https://www.sciencedirect.com/science/article/pii/S0092867418301545 ↩
-
Codella et al., “Skin lesion analysis toward melanoma detection: A challenge at the 2017 International symposium on biomedical imaging (ISBI), hosted by the international skin imaging collaboration (ISIC)”, ISBI, 2018: https://arxiv.org/abs/1710.05006 ↩