Combined tract segmentation and orientation mapping for bundle-specific tractography
Highlights
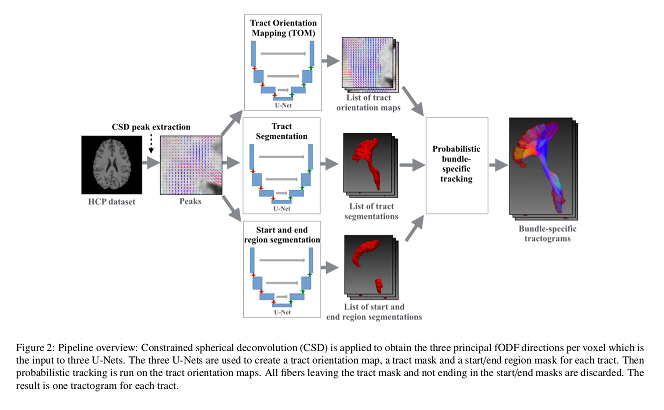
Authors propose to learn the track orientation mapping (TOM) and combine it with the bundle outline and connected regions to perform bundle-specific tractography.
The work builds up on a number of other works by the same group, including the TractSeg method. An early version of it was reviewed here.
Introduction
A number of approaches have been proposed to filter the false positives that current tractography techniques produce. These approaches can be grouped into two categories:
- Clustering-based approaches: streamlines are filtered based on their spatial relation to cortical or other anatomically defined regions.
- Region-of-interest-based approaches: group and select streamlines by measuring intra- and inter-subject streamline similarities.
Authors propose to combine some of their earlier works, notably TOM and TractSeg, and to add another bundle start and end filtering network to overcome some of the limitations that these methods present individually.
Methods
Authors use their TOM method to obtain the list of orientation maps (a local mean streamline orientation of the track). TractSeg is used for white matter bundle segmentation. Finally, another convolutional neural network is used to segment the bundle start and end regions, which helps filtering tracks that end prematurely or do not connect the regions corresponding to that bundle.
A probabilistic approach is used to obtain the tractography result using the output of the three convolutional networks.
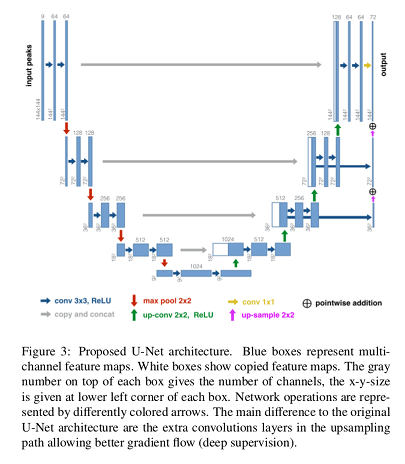
Authors propose to use the same 2D U-Net-based architecture for the three networks. Random orientation sampling on the axial, coronal, and sagittal directions is performed during training for bundle and region segmentation. At inference time, the mean value of the predictions is used.
Losses:
- Cosine similarity + squared l2-norm for TOM.
- Binary cross-entropy for the segmentation networks.
The network input is composed of the fODFs peaks computed at each voxel. The three principal fiber directions are kept for each voxel using the multi-tissue multi-shell constrained spherical deconvolution (CSD) method.
Authors used bundle-specific thresholds to improve results (especially on bundles that are harder to track) to convert the computed probability maps to binary maps.
Data augmentation was carried out applying:
- Affine transformation
- Elastic transformation
- Scaling
- Resampling
- Gaussian noise
Data
Training was performed on the 105-subject, 72 bundle per subject HCP dataset.
Testing was performed on 17 datasets, including TRACED, Sherbrooke_3shell, HCP, ADNI and OASIS. This means that the data used by the authors was acquired with different scanners, different field strengths, different acquisition protocols, different image qualities, and on brains with different diseases.
Results
They used 3 widely used methods and 4 in-house methods (used earlier in their TractSeg method) as their baselines:
- TractQuerier
- RecoBundles
- TRACULA
- Atlas registration
- Multiple mask registration
- Peak atlas
- Best original peak
Authors used 5-fold cross validation to select the parameters.
Metrics:
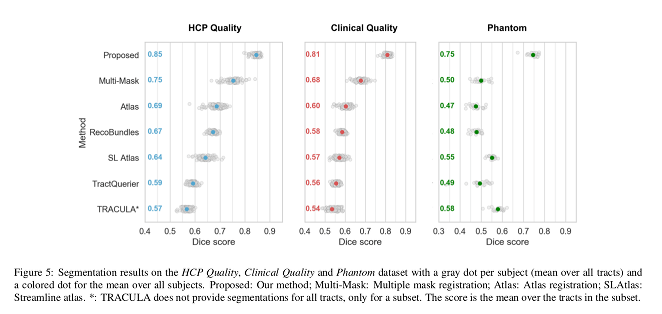
- Segmentation performance: Dice score.
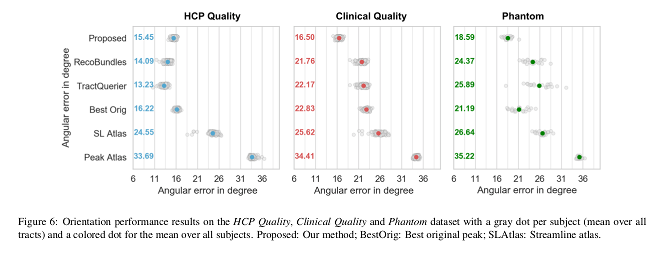
- Orientation performance: voxel-wise angular error.
Their method is faster and produces more accurate bundle-specific tractograms.
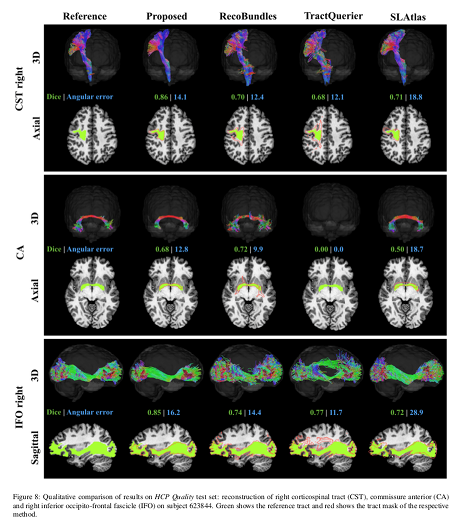
Conclusions
The presented method outperforms the baselines, providing more accurate bundle segmentations and tractograms, and is less affected by the reduction in resolution in the clinical quality data.
It also generalizes well to unseen datasets and pathologies.