Direct White Matter Bundle Segmentation using Stacked U-Nets
Summary
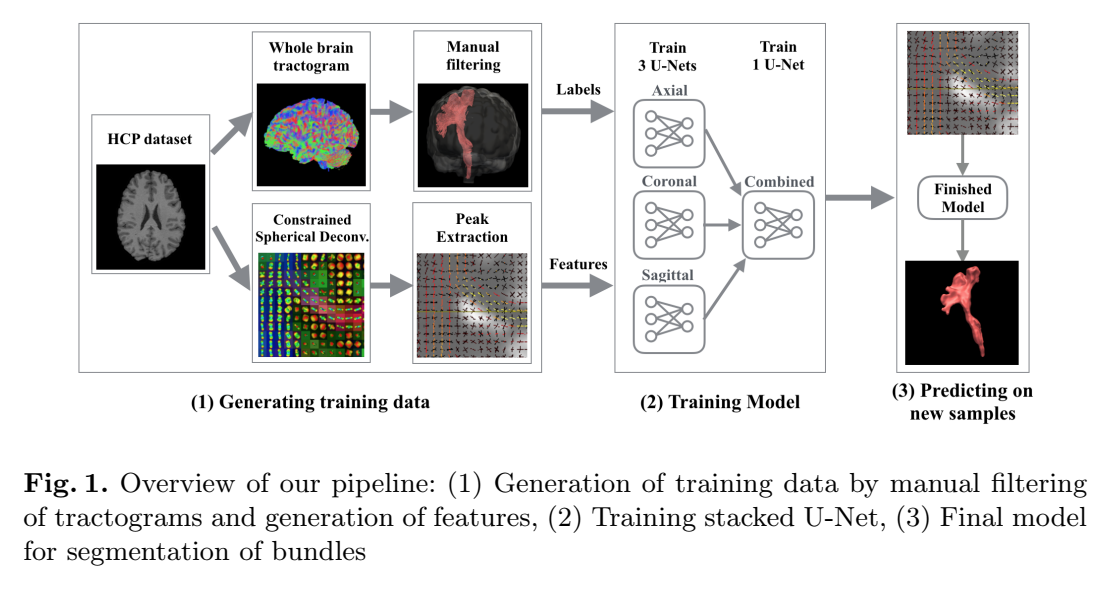
The authors propose a U-Net based approach for direct bundle segmentation. This means that tractography is effectively bypassed, and no streamlines are needed to predict a bundle’s probability map. The raw diffusion signal (or SH coeffs) is too large to fit in memory, therefore the images are processed and 3 fODFs peaks are extracted per voxel (using MRtrix CSD + peak extraction). Input images thus have 9 channels instead of between 50 and 200.
The model is used for a binary classification task; the final prediction is a 3D probability map, where each voxel’s value is its probability of belonging to the bundle of interest.
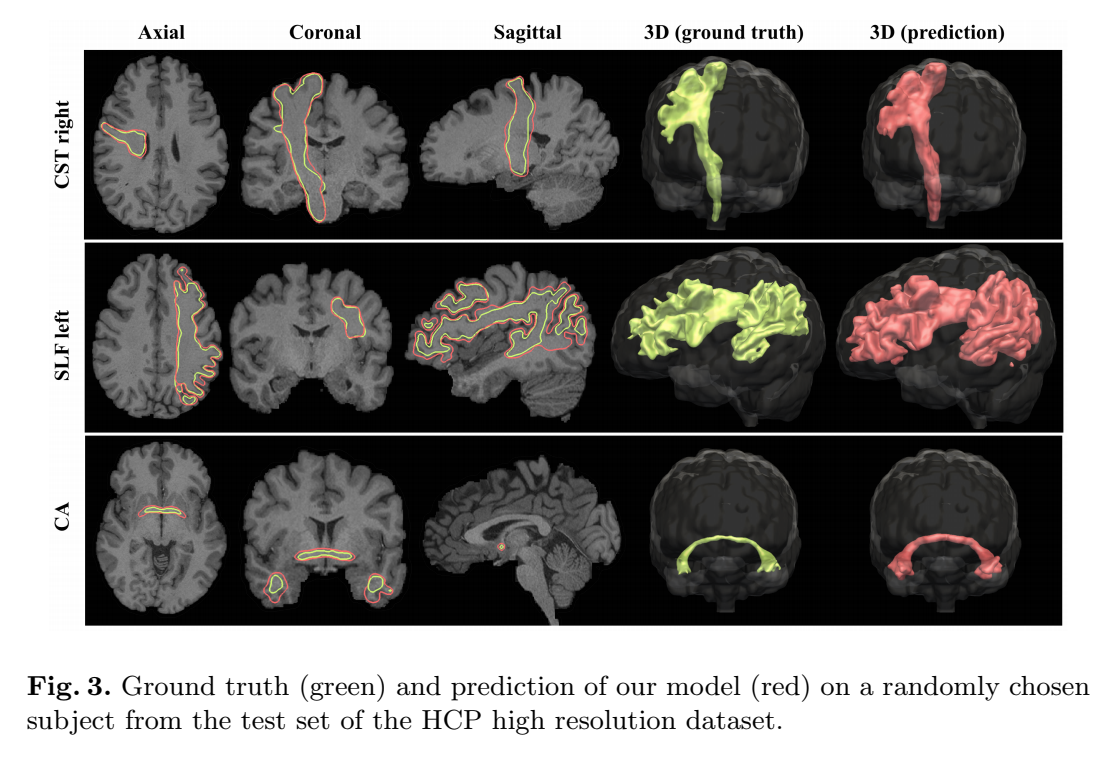
Training data is obtained by 1) whole-brain tracking, 2) valid streamlines segmentation for the chosen bundle, then 3) generating a binary coverage map.
Model
- Three U-Nets trained independently (one per axis) = 3 predictions per voxel
- Fourth U-Net takes the 3 predictions combined as a 3-channel input image (3D segmentation? Unclear…)
- Binary classification target (bundle-specific model)
Experiments and Results
Datasets
- 3 tracts/bundles : SLF left (medium), CST right (hard), CA (very hard)
- 30 HCP subjects, high resolution: 1.25mm isotropic, 270 directions, b1000+b2000+b3000, “manually” segmented for the 3 bundles (using 1 million multi-shell CSD prob streamlines + MRtrix segmentation pipeline using WM atlas ROIs)
- HCP low resolution: HCP high resolution resampled to 2mm isotropic, b1000, 32 directions evenly distributed on the sphere
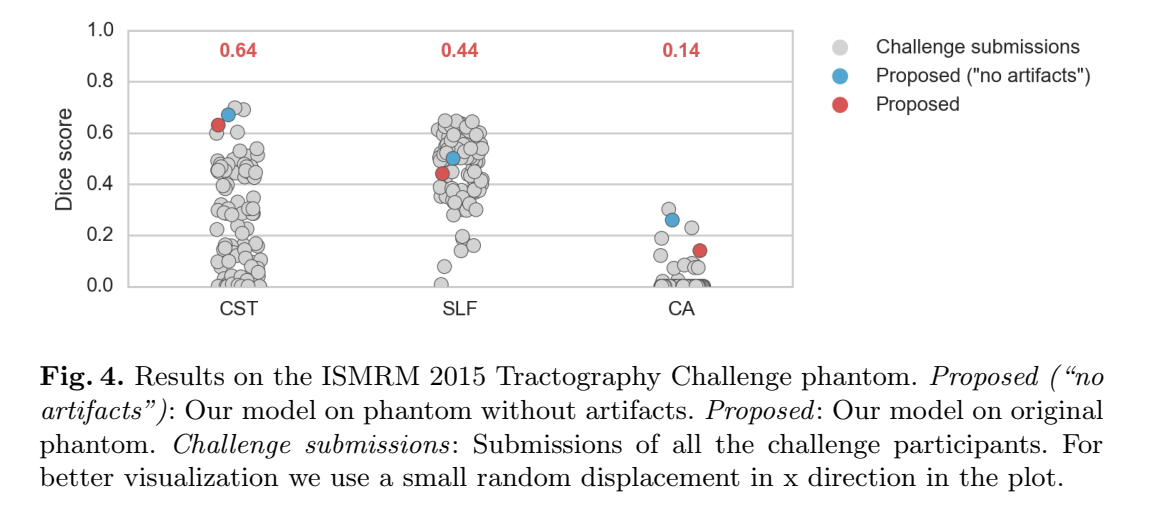
- 2015 ISMRM tractography challenge: phantom with simulated artefacts and noise + MRtrix denoising and correction, known ground truth
- Same as 3, but “perfect” phantom without simulated artifacts/noise
Experiments
- Training and testing on HCP high res
- Training and testing on HCP low res
- Training on HCP low res and testing on ISMRM challenge
Baselines
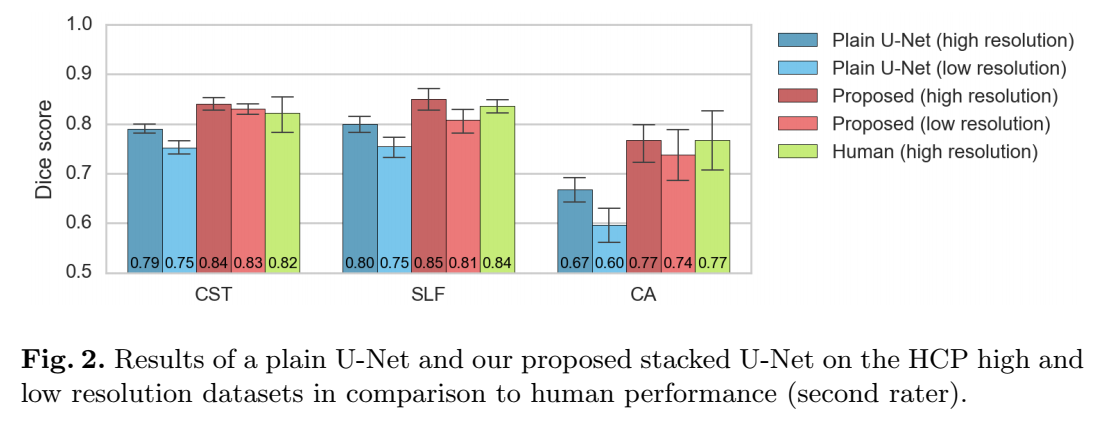
- Human performance: Automatic segmentation + one human segmentation