Joint Learning of Brain Lesion and Anatomy Segmentation from Heterogeneous Datasets
Highlights
-
Authors propose to train a single CNN model to predict brain tissue and lesion segmentations using heterogeneous datasets labeled independently.
-
A novel adaptive cross-entropy loss function is defined to overcome label contradiction issues that may arise in this task. Their trick is to reinterpret the meaning assigned to the lesion background label (in blue) as any label that is not lesion and modify the loss function accordingly.
-
Benchmarking is done against a naive model and a multi-network approach.
Introduction
During the last years, convolutional neural networks (CNNs) proved to be highly accurate to perform medical image segmentation.
Even if most publicly available datasets provide image annotations for single tasks, in practice it is usually desirable to train single models which can learn to perform multiple segmentation tasks simultaneously.
Learning to segment multiple structures from heterogeneous datasets is challenging since labels coming from different datasets may contradict each other and misleading the training process.
Authors work labels brain lesion and anatomy segmentation learned from heterogeneous datasets (i.e. each dataset is annotated for a single task).
Related work includes 1 and 2. In 1 semantic full scene labeling in outdoor images coming from different datasets with heterogeneous labels is done. Their loss function is data-selective. In 2, reviewed in here, multiple labels may be assigned to a single voxel.
Methods
Disjoint labelsets are assumed except for the background label. Problematic areas are those for which the original lesion datasets indicate background label, while they should be annotated as actual tissue labels.
A single label is assigned to every voxel.
Authors propose a new loss function called Adaptive Cross Entropy (ACE) to overcome the contradictory label problem. The loss is defined differently depending on the structures of interest under consideration: in the lesion dataset, the meaning assigned to the background label is a virtual class “any label that is not lesion”) and modify the loss function accordingly. The proposed adaptive cross-entropy is show in Figure 2.
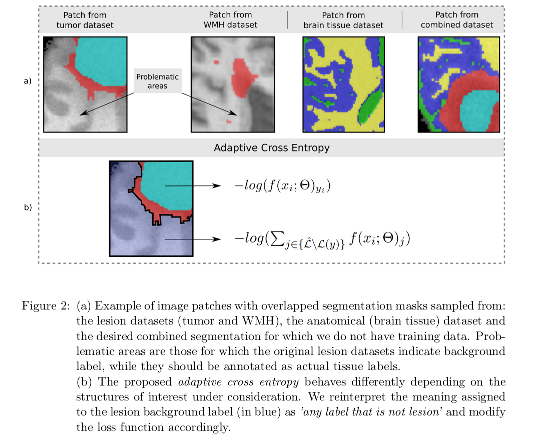
- When the label is not in conflict, minimizing the entropy is equivalent to maximizing the score for the correct class.
- When dealing with a voxel whose ground truth is lesion background (i.e. it may be any healthy brain tissue label), the model tends to maximize the probability for all non-lesion classes (i.e. the virtual “any label that is not lesion” class).
The CNN used by the authors is a standard U-Net.
Data
Brain Magnetic Resonance Images (MRI) from a number of publicly available datasets or challenges containing different weighting and sequences (T1, T1g, T2 and FLAIR):
- MRBrainS13 Challenge
- The WMH Segmentation Challenge (WMH 2017)
- MRBrains18
- Brainweb 3
- BRATS2012
- Tumorsim 4
Authors consider the joint learning of brain tissue segmentation and two separate type of lesions: brain tissue vs. White Matter Hyperintensity (WMH), and brain tissue vs. tumor and edema lesions.
Given the lack of datasets with simultaneous annotations for brain tumors and tissues, synthetic and simulated images are used.
Benchmarking
Baselines:
- Naive model: a single U-Net is trained by minimizing standard loss functions (Dice and cross-entropy).
- Multi-network baseline (Multi-UNet): multiple independent networks trained on each task using a categorical cross-entropy loss. Segmentation results are then combined following the criterion that in case of brain lesion and tissue segmentation lesion labels prevail.
Results
The proposed ACE makes it possible to train a single model for joint learning of brain lesion and anatomy from heterogeneous datasets, achieving equivalent performance to that of Multi-UNet.
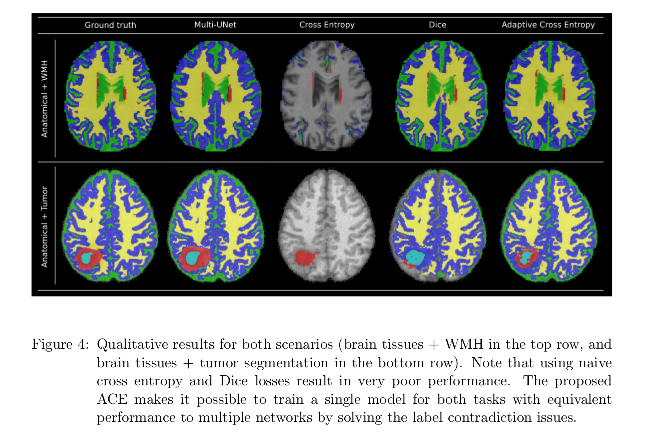
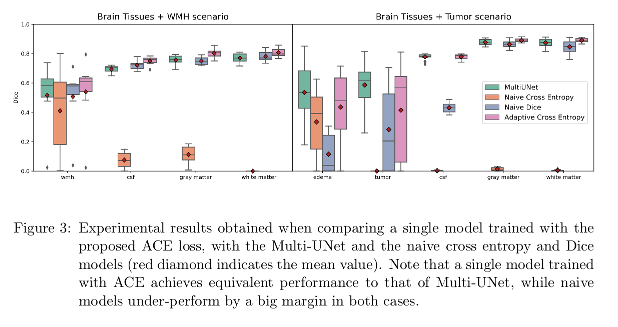
Conclusions
A single CNN model to perform joint brain lesion and anatomy segmentation using a new loss function (Adaptive Cross-Entropy) is presented. Results are competitive with the baselines.
Compared to the Multi-UNet approach, the presented approach has the limitation of the fixed number of input channels (only use those sequences available in both anatomy and lesion datasets can be used by their model). Multi-UNet, in turn is trained on all available modalities for a given task.
Also, the comparison with the single UNet is not fair since it was trained using only T1 images.
Domain adaptation to overcome multi-domain problems (different sequences give rise to different image intensities), extension to the segmentation of other sub-cortical structures and other CNN architectures are mentioned as a future work.
References
-
Damien Fourure, Rémi Emonet, Elisa Fromont, Damien Muselet, Natalia Neverova, Alain Trmeau, and Christian Wolf. Multi-task, multi-domain learning: Application to semantic segmentation and pose regression. Neurocomputing, 251:68-80, 2017. ISSN 0925-2312. doi: https://doi.org/10.1016/j.neucom.2017.04.014. ↩ ↩2
-
Martin Rajchl, Nick Pawlowski, Daniel Rueckert, Paul M Matthews, and Ben Ben Glocker. Neuronet: Fast and robust reproduction of multiple brain image segmentation pipelines. International Conference on Medical Imaging with Deep Learning (MIDL) 2018. ↩ ↩2
-
Chris A. Cocosco, Vasken Kollokian, Remi K.-S. Kwan, G. Bruce Pike, and Alan C. Evans. Brainweb: Online interface to a 3D MRI simulated brain database. NeuroImage, 5:425, 1997. ↩
-
Marcel Prastawa, Elizabeth Bullitt, and Guido Gerig. Simulation of brain tumors in MR images for evaluation of segmentation efficacy. Medical image analysis, 13(2):297–311, 2009. ↩