Deep Multi-Structural Shape Analysis: Application to Neuroanatomy
Highlights
- Authors propose a deep neural network for supervised learning on neuroanatomical shapes.
- Their method works directly on point clouds without requiring a prior triangulation step; the network learns the optimal representation of the shape descriptors.
- The method is applied to: (i) prediction of Alzheimer’s Disease and mild cognitive impairment (multi-class classification); (ii) regression on brain age.
Introduction
Shape analysis of anatomical structures is of core importance for many tasks in medical imaging.
The use of deep networks in medical shape analysis is still largely unexplored; among others, because point cloud representations do not possess an underlying Euclidean or grid-like structure.
Traditional approaches rely on computing pre-defined shape features. The variational auto-encoder architecture proposed by Shakeri et al.1 does not perform an end-to-end training, relies on computing mesh correspondences and focuses on a single structure.
Authors introduce a method for medical shape data modelling based on PointNet, a deep neural network architecture which operates directly on a point cloud and predicts a label in an end-to-end fashion. Their method, named Multi- Structure PointNet (MSPNet), is able to simultaneously predict a label given the shape of multiple structures.
Methods
- Two stages
- Extraction of point clouds representing the anatomy of different structures from medical images: segmentation and uniform sampling (i.e. all structures have the same number of points).
- Multi-Structure PointNet (MSPNet). Multi-branch structure, where each branch processes the point clouds belonging to a single structure. Steps: 1) point cloud alignment using a transformation network; 2) feature extraction; 3) feature alignment with a second transformation net; 4) dropout; 5) prediction.
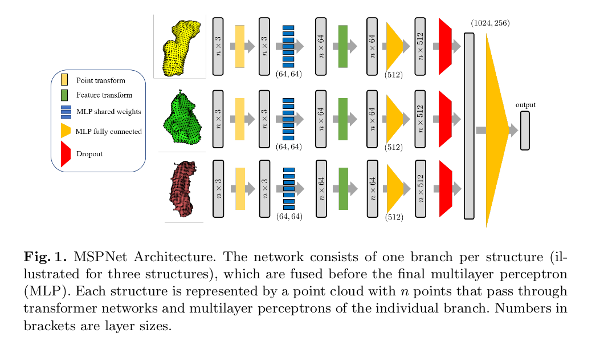
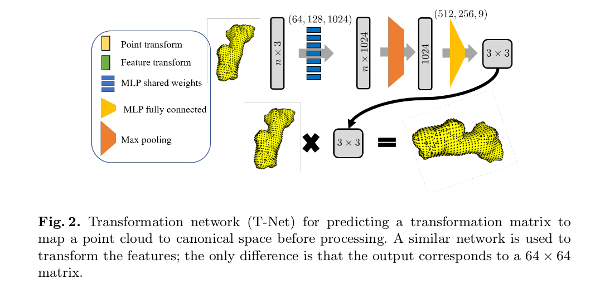
The transformation network (T-Net) is a multilayer perceptron (MLP) estimating a rigid transformation into a canonical space.
Weights are shared among points in the feature extraction network.
Data augmentation is done applying random rigid transformation to training points.
Both for the Alzheimer’s Disease (AD) vs. mild cognitive impairment classification (MCI), and the age classification task the shape of the left and right hippocampus and the left and right lateral ventricles are used.
For the age classification task, two experiments are performed: one using only the healthy controls (HC), and another one including patients diagnosed with MCI and AD.
Results
The Alzheimer’s Disease Neuroimaging Initiative (ADNI) database MRI T1-weighed 7,974 images (2,423 healthy subjects; 978 Alzheimer’s Disease, and 4,625 mild cognitive impairment) are used.
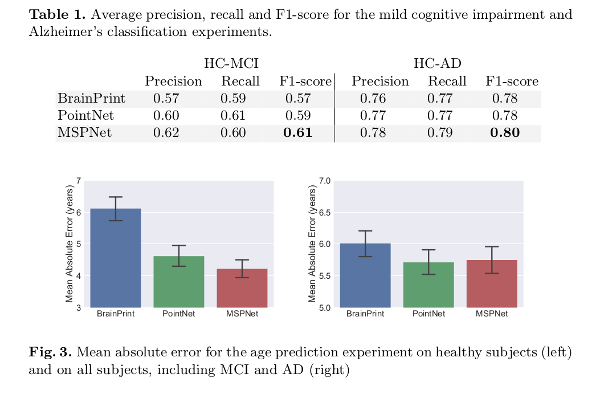
Results are compared to the standard PointNet architecture and spectral shape descriptors in BrainPrint2.
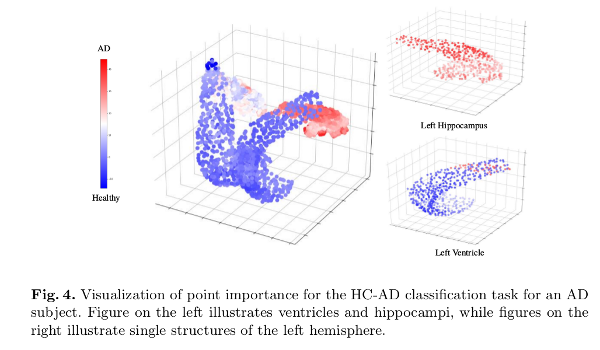
Additionally, a point importance experiment is performed using the occlusion method.
Discussion
MSPNet outperforms the compared methods in all tasks but the age prediction task when using all subjects (where PointNet showed a better performance).
Comments
- The segmentation process could be part of the deep learning process.
- The network requires as many branches as structures, which need to be known beforehand. Also, just those knowing to affect the result are used, which assumes an a priori knowledge.
References
-
Mahsa Shakeri, Herve Lombaert, Shashank Tripathi, and Samuel Kadoury. Deep Spectral-Based Shape Features for Alzheimer’s Disease Classification. Proceedings of International Workshop on Spectral and Shape Analysis in Medical Imaging (SeSAMI). Medical Image Computing and Computer Assisted Interventions (MICCAI), 15-24. 2016. ↩
-
Christian Wachinger, Polina Golland, William Kremen, Bruce Fischl, and Martin Reuter. BrainPrint: A discriminative characterization of brain morphology. Neuroimage 109, 232-248 (2015). ↩