MedTrinity-25M: A Large-scale Multimodal Dataset with Multigranular Annotations for Medicine
Highlights
This paper introduces the largest dataset designed to support multimodal medical AI tasks. It covers over 25 million images across 10 modalities with detailed annotations for more than 65 diseases. The dataset includes global textual information like disease type, modality, and region-specific descriptions, as well as local annotations for regions of interest (ROIs) such as bounding boxes and segmentation masks.
Key Features of MedTrinity-25M:
- Automated Data Construction: Built using an automated pipeline that scales up multimodal data by generating multigranular annotations from unpaired images without relying on text descriptions.
- Dataset Composition: Data from over 90 sources, preprocessed with expert models identifying ROIs related to abnormal regions. The data includes multigranular visual and textual annotations.
- Applications: MedTrinity-25M has proven very effective for visual Q&A tasks.
The most compelling aspects of this paper are (1) the dataset’s creation, curation, and automatic annotation process, and (2) its role in enabling multimodal large language models (MLLMs) to achieve state-of-the-art (SOTA) results on three VQ&A datasets.
Dataset Creation and Automatic Annotation
The MedTrinity-25M dataset is composed of triplets: ( Image, Region of Interest (ROI), Description). The dataset creation process is illustrated in Figure 2.
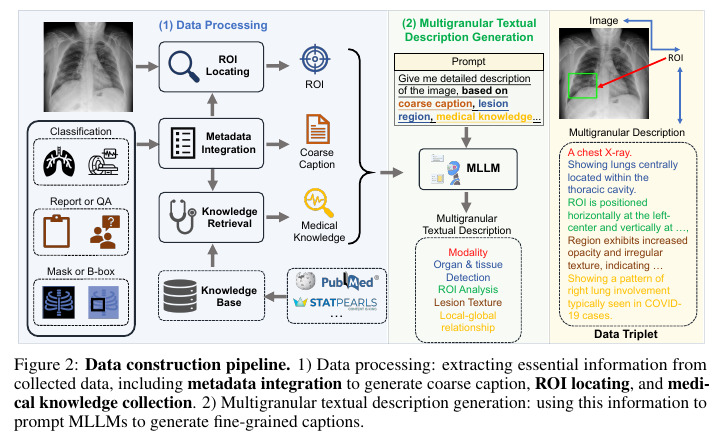
The Automated Data Construction process generates large-scale, multigranular annotations for medical images. Key steps include:
1. Data Collection and Preprocessing
- Assembled from over 90 online resources, including TCIA, Kaggle, Zenodo, and Synapse.
- Includes medical images with varying levels of existing annotations, such as segmentation masks, lesion bounding boxes, or disease types, often lacking detailed textual descriptions.
- Preprocessing involved:
- Identification of Regions of Interest (ROIs): Using domain-specific expert models to locate abnormalities within the images.
- Metadata Integration: Extracting and integrating metadata to generate coarse captions providing fundamental information about each image, including modality, organ labels, and disease types.
- Medical Knowledge: To enrich reports with specialized medical terminology and professional expression, they built a medical knowledge database following the MedRAG approach1. Retrieval-Augmented Generation (RAG) ensures that the generated text remains contextually relevant, reducing hallucinations by providing the LLM with relevant information. Medical text was collected from PubMed for biomedical knowledge, StatPearls for clinical decision support, and medical textbooks for domain-specific knowledge.
2. Generation of Multigranular Annotations
The automated pipeline uses Multimodal Large Language Models (MLLMs) to generate detailed visual and textual annotations without expert input. Using a prompt template, the MLLM is prompted with annotations that include Global Information such as disease/lesion type, modality, and inter-regional relationships, and Local Information including detailed descriptions for ROIs, such as bounding boxes, segmentation masks, and specific textual descriptions. These annotations help create comprehensive image-ROI-description triplets.
3. Quality Validation
The generated multigranular descriptions are validated against human annotations to assess accuracy and alignment, comparing structured descriptions generated by the MLLMs with human-generated text to ensure accuracy and comprehensiveness.
An example of this automatic annotation is illustrated here:
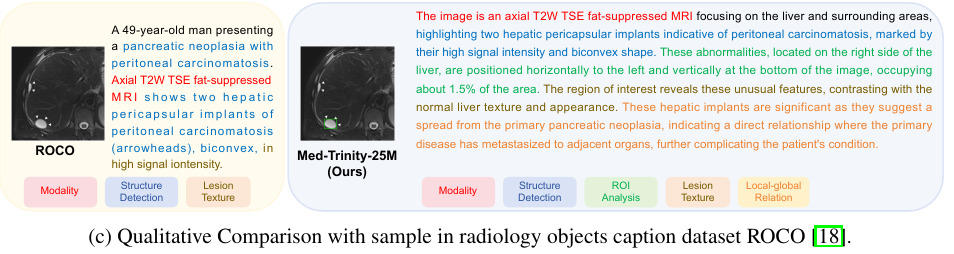
Visual Q&A
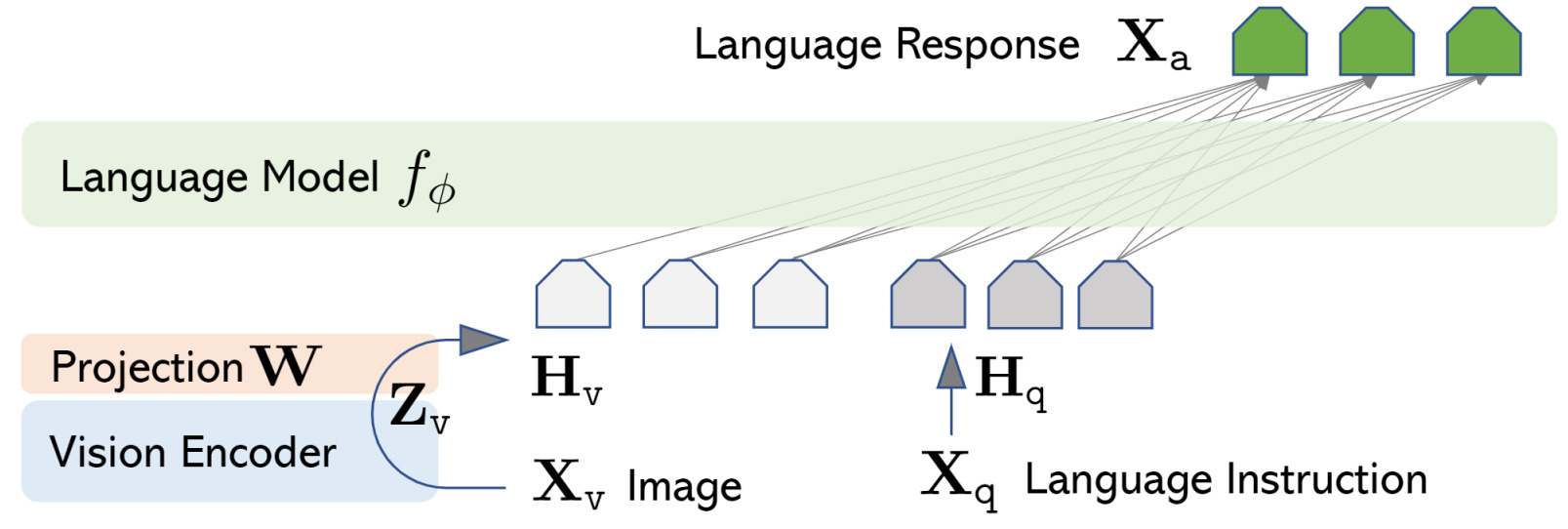
To demonstrate the power of their dataset, the authors implemented a multimodal vision LLM called Large Language and Vision Assistant (LLaVA)2. LLaVA combines a vision encoder with Vicuna3 for general-purpose visual and language understanding. The core idea behind LLaVA is illustrated in the following figure where \(f_\phi\) is Vicuna:
Vicuna3 is an open-source Large Language Model (LLM) based on Meta’s LLaMA architecture. It excels in generating conversational, human-like text responses. Key features include:
- LLaMA-based: Built on the efficient LLaMA model, offering strong performance with fewer resources.
- Open-Source: Accessible for researchers and developers to fine-tune and deploy.
- Conversational Focus: Optimized for dialogue, ideal for chatbots and virtual assistants.
- Efficient: Delivers coherent responses while being resource-efficient.
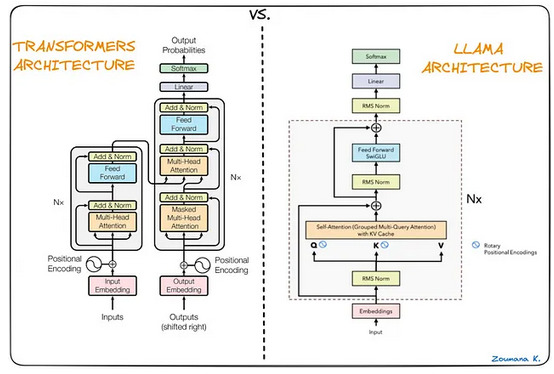
As a reminder, LLaMA4 is Meta’s LLM, with the following main characteristics:
- Model Variants: Four versions with 7B, 13B, 32.5B, and 65.2B parameters, featuring high-dimensional learned embeddings.
- Attention Enhancements: Grouped multi-query attention and KV caching for faster, efficient inference.
- Normalization: Uses RMS Normalization for stable training.
- Positional Encoding: Implements Rotary Positional Embedding (RoPE) for dynamic token positioning.
- Activation Function: Employs SwiGLU in feed-forward layers for improved performance.
These modifications make LLaMA a highly efficient and powerful language model.
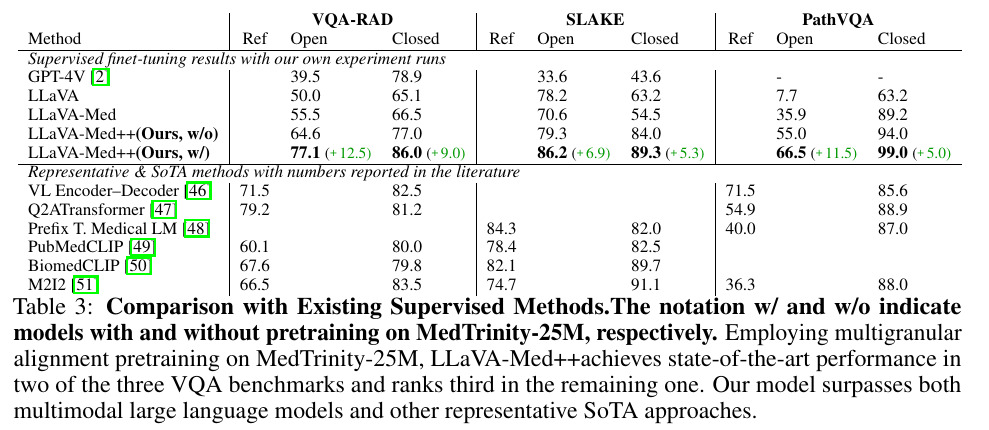
Results and Dataset
The results obtained on three large medical Visual Q&A datasets reveal that LLaVA-Med++ is SOTA as of today.
Everything about MedTrinity-25M is available here.
References
-
G. Xiong, Q. Jin, Z. Lu, and A. Zhang. Benchmarking retrieval-augmented generation for medicine. arXiv preprint arXiv:2402.13178, 2024. ↩
-
H Liu, C Li, Q Wu, YJ Lee Visual Instruction Tuning, NeurIPS 2023 ↩
-
W-L Chiang, Z Li, Z Lin, Y Sheng, Z Wu, H Zhang, L Zheng, S Zhuang, Y Zhuang, J.E. Gonzalez, I Stoica, and E.P. Xing. Vicuna: An open-source chatbot impressing GPT-4 with 90% ChatGPT quality ↩ ↩2
-
https://medium.com/@pranjalkhadka/llama-explained-a70e71e706e9 ↩