Towards Segment Anything Model (SAM) for Medical Image Segmentation: A Survey
Introduction
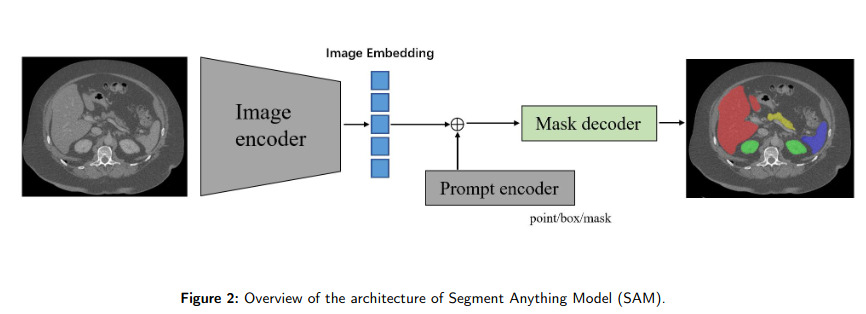
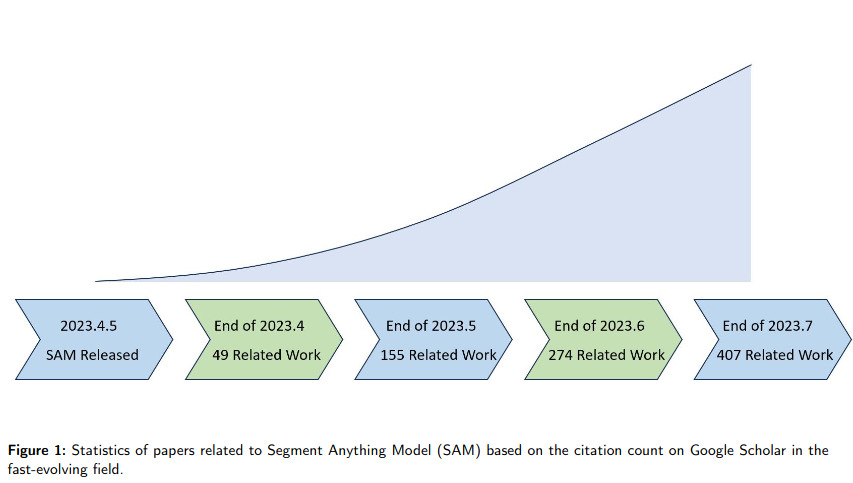
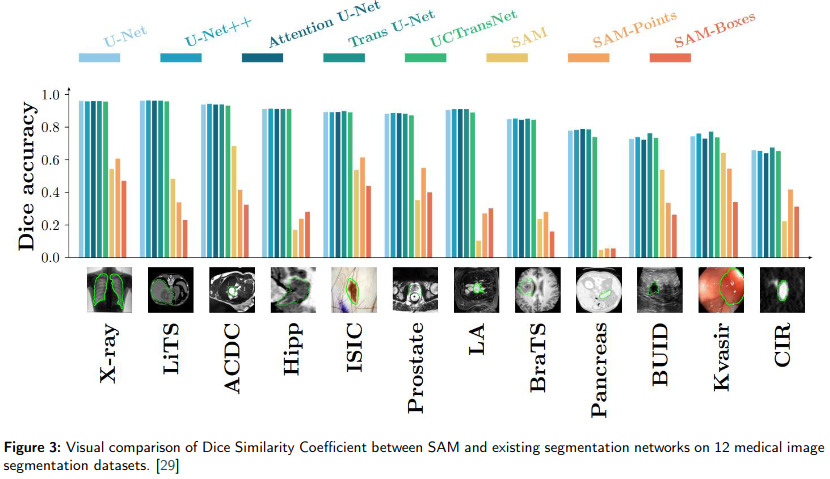
SAM (Segment Anything Model) is a foundation model for image segmentation (c.f. Fig 2). It has shown promise in natural image processing and image generation due to its flexibility in prompting (c.f. Fig 1). The main takeaway of this paper is that SAM’s applicability in medical image segmentation, which differs significantly from natural images, remains uncertain (c.f. Fig 3). This paper reviews efforts to adapt SAM to medical image segmentation tasks, including empirical benchmarking and methodological adaptations, and discusses potential future directions.
SAM’s Performance in Medical Image Segmentation
The paper examines SAM’s effectiveness in 12 medical image segmentation tasks, such as
- Pathology Image Segmentation:
- Tumor segmentation.
- Non-tumor tissue segmentation.
- Cell nuclei segmentation on whole slide imaging (WSI).
- Liver Tumor Segmentation from Contrast-Enhanced Computed Tomography (CECT) Volumes.
- Polyps Segmentation from Colonoscopy Images.
- Brain MRI Segmentation: - Comparing SAM with the Brain Extraction Tool (BET) for brain extraction and segmentation.
- Abdominal CT Organ Segmentation:
- Evaluating zero-shot capabilities of SAM for abdominal organ segmentation.
- Endoscopic Surgical Instrument Segmentation:
- Performance evaluation of SAM on datasets for endoscopic surgical instrument segmentation.
- Multi-Dataset Evaluations:
- Comprehensive studies evaluating SAM’s accuracy on various medical image segmentation datasets covering different organs (e.g., brain, breast, chest, lung, liver, bowel, pancreas, prostate), image modalities (e.g., 2D X-ray, histology, endoscopy, 3D MRI, CT), and health conditions (normal, lesioned).
Adapting SAM for Medical Image Segmentation:
The document outlines several ways to better adapt the Segment Anything Model (SAM) to medical image segmentation.
Fine-Tuning SAM on Medical Datasets: Partial fine-tuning of SAM, particularly its image encoder, prompt encoder, or mask decoder, enhances its performance on medical images. Examples include skin cancer and polyp segmentation.
Extending SAM’s Usability for Medical Images: Adapting SAM for typical medical image formats and viewers. AutoSAM and All-in-SAM are initiatives for automatic segmentation and label-efficient fine-tuning, respectively.
Enhancing SAM’s Robustness to Prompts: Research has focused on making SAM less dependent on manual prompts by decoupling its mask decoder into prompt-relevant and prompt-invariant modules, and by using multiple box prompts for better accuracy.
Input Augmentation with SAM: To overcome SAM’s limitations in direct medical segmentation, researchers are augmenting raw images with SAM-generated masks to improve segmentation results.
Future Directions and Challenges:
The paper outlines challenges in building large-scale medical datasets, integrating clinical domain knowledge, adapting SAM from 2D to 3D medical images, and reducing annotation costs for medical image segmentation. It emphasizes the importance of these areas for the future development of foundation models for medical image segmentation.