Fully Automated, Quality-Controlled Cardiac Analysis From CMR
Introduction
Clinical use of deep learning algorithms in cardiac image segmentation analysis is limited due to unavoidable issues with image quality, artifacts and anatomical variation.
The objective was to develop robust quality control measures in order to facilitate translation of deep learning methods of cardiac magnetic resonance (CMR) image analysis into clinical pratice.
Methods
Pipeline
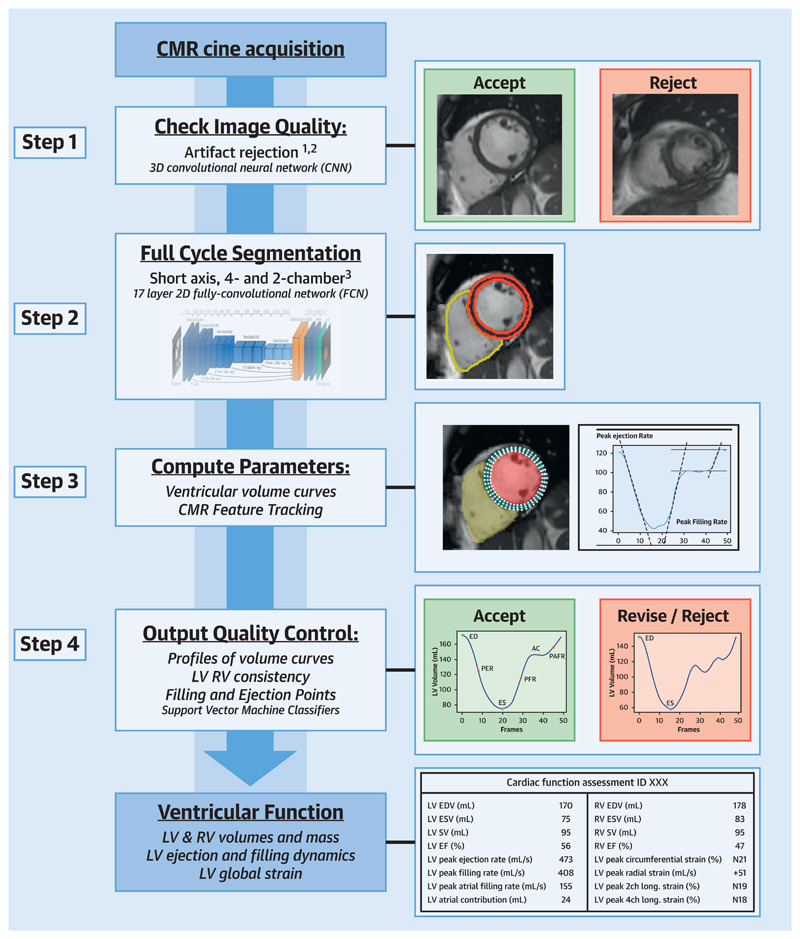
Step 1: Pre-analysis image QC (QC1)
The first quality control section aims to detect and reject bad quality images based on two categories: motion artifacts and erroneous planning.
In both cases ROI extraction is used to deal with variable image size, in order to avoid issues with mofication of image quality in the processing steps. Input size was 80x80 pixels and 64x64 pixels for motion artifact detection and erroneous planning respectively.
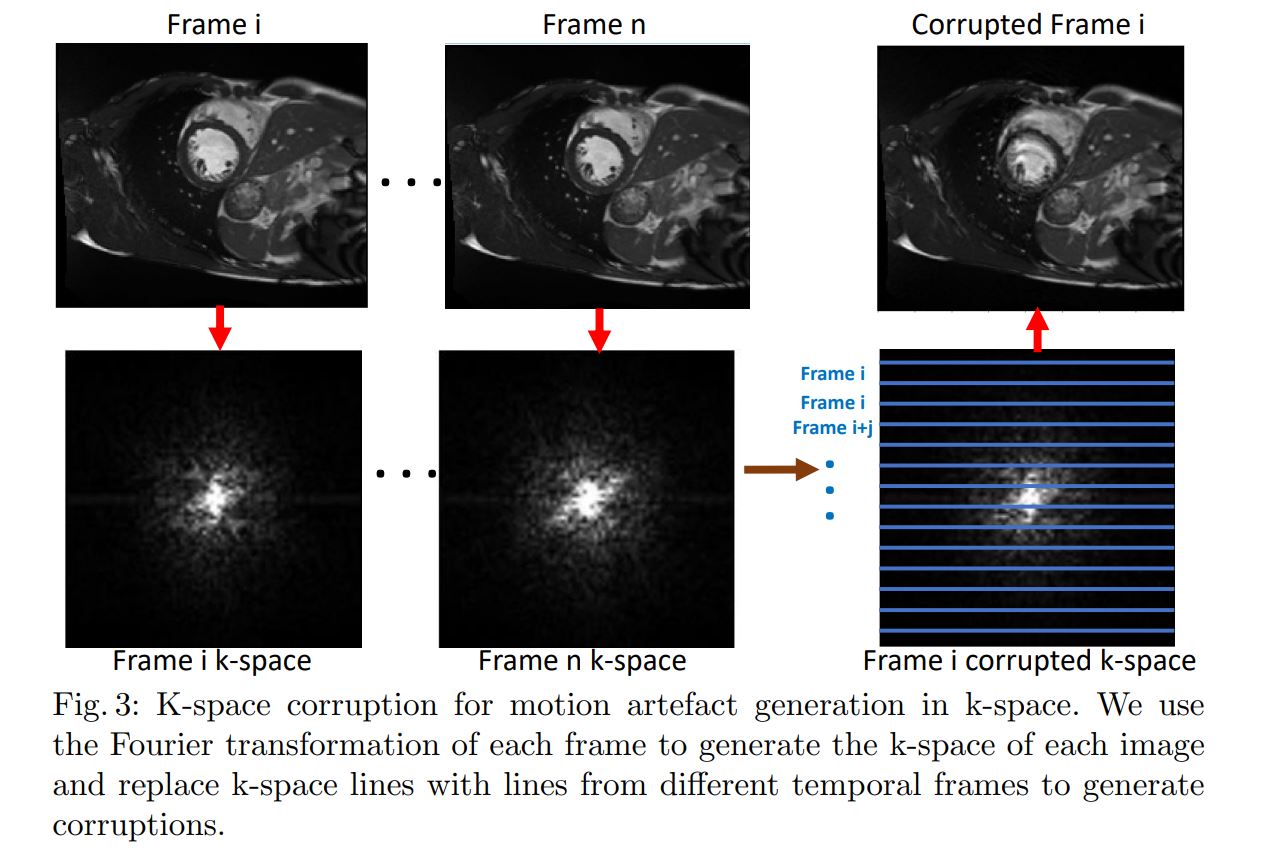
Motion artifact detection
- 2D CNN with LSTM layer
- k-space corruption for data augmentation to provide balanced classes. In the fourrier space, change some lines from one image slice with lines from other slices 1.
Detection of erroneous planning of 4 chamber view
- Classification of presence of left ventricular outflow tract (LVOT) in images (246 CMR images from UK Biobank).
- 5 layer, 2D CNN (LeNet architecture) outperformed other classifiers such as K-Nearest Neigbours, SVM, Random Forest, etc. 2
Step 2: Image segmentation
Segmentation of LV, RV and LV myocardium in all frames. Trained on manually annotated data. Dataset contained 3975 subjects containing subjects with cardiac diseases and healthy subjects.
Step 3: Parameter calculation
Steps used in parameter calculations:
- Align short and long axis imaging stacks
- Calculate LV and RV volume curves and LV mass (LVM)
- Calculate metrics from volume curves (EDV, ESV, peak ejection rate, etc.)
- Feature tracking with end-diastolic LV wall as ROI. Using Medical Image Registration ToolKit (https://mirtk.github.io/).
- More metrics from feature tracking. Included global circumferential strain, radial strain and longitudinal strain.
Step 4: Post-analysis QC (QC2)
QC2 contains 3 parts in order to flag cases for clinician review.
- Validation of orientation of images and missing slices
- Inspection of output parameters; flag suspicious exams.
- Difference >10% on LV and RV SV
- Difference of volume between phases
- SVM classification for abdnomalities in volume and strain curves.
- Data used for training these classifiers was from healthy subjects and subjects with cardiomyopathy from UK Biobank, annotated by an expert.
Results and Discussion
Importance of QC1 lies in the significance of the impact of motion artifacts and off-axis planning on the computed metrics. These issues will not be recognisable easily in post-analysis QC, therefore are best flagged before.
QC2 relies on anatomical and clinical knowledge to identify abnormalities. According to the authors, this can extend to detection of cases not anticipated in the development of the algorithms.
Authors believe that including an uncertainty score in the segmentation networks could be beneficial to the CMR analysis.
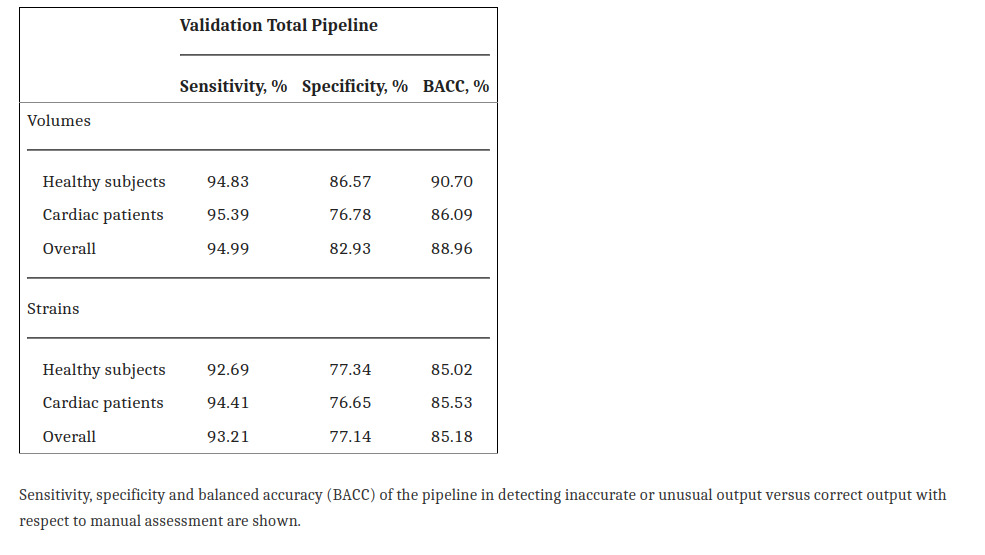
Pipeline validation done with comparisons with expert review showed that the QC steps correctly detected erroneous cases.
Rate of flagged CMRs | | Pipeline | Clinicial review | |—|—|—| | Healthy patients| 26% | 15.2% | | Cardiac patients | 32% | 11% |
It seems however that the pipeline may systematically remove unhealthy subjects based on medical condition rather than image/metric quality.
Sensitivity to errors in volume curves and strain was high. However, the specificity shows that it is very sensitive to false positive cases (flags valid sequences as erroneous)
Conclusions
Quality control measures allow fully autonomous processing of CMR exams, while flagging possible problematic cases for clinician review.
The pipeline can facilitate the creation of a large set of reference values for characterization of cardiac function. The pipeline allows for much shorter computation time compared to commercial software for semi-automated segmentation and feature tracking.
References
-
Oksuz I, Ruijsink B, Puyol-Anton E, et al. Deep learning using K-space based data augmentation for automated cardiac MR motion artefact detection. In: Medical Image Computing and Computer-Assisted Intervention - MICCAI 2018. Lecture Notes in Computer Science. Springer, 2018;11070:250–8. ↩
-
Oksuz I, Ruijsink B, Puyol-Anton E, et al. Automatic left ventricular outflow tract classification for accurate cardiac MR planning. IEEE 15th Int Symp Biomed Imaging 2018:462–5 ↩