TractoFormer: A Novel Fiber-level Whole Brain Tractography Analysis Framework Using Spectral Embedding and Vision Transformers
Highlights
- TractoEmbedding can generate images representative of fiber populations
- TractoFormer can use these images to perform discriminative tasks
Introduction
Defining a good data representation of tractography for machine learning is still an open challenge, especially at the fiber level.
Another challenge in machine learning for tractography analysis is the limited sample size (number of subjects) of many dMRI datasets. Developing data augmentation methods to increase sample size is a known challenge in structural connectivity research.
Methods
Data
Two datasets are considered:
- 100 subjects from the Human Connectome Project
- 150 subjects (103 healthy, 47 schizophrenia patients) from the Consortium for Neuropsychiatric Phenomics dataset.
Whole-brain tractograms (1M streamlines per subject) are generated using two-tensor unscented Kalman Filter tractography (from SlicerDMRI). All tractograms are coregistered.
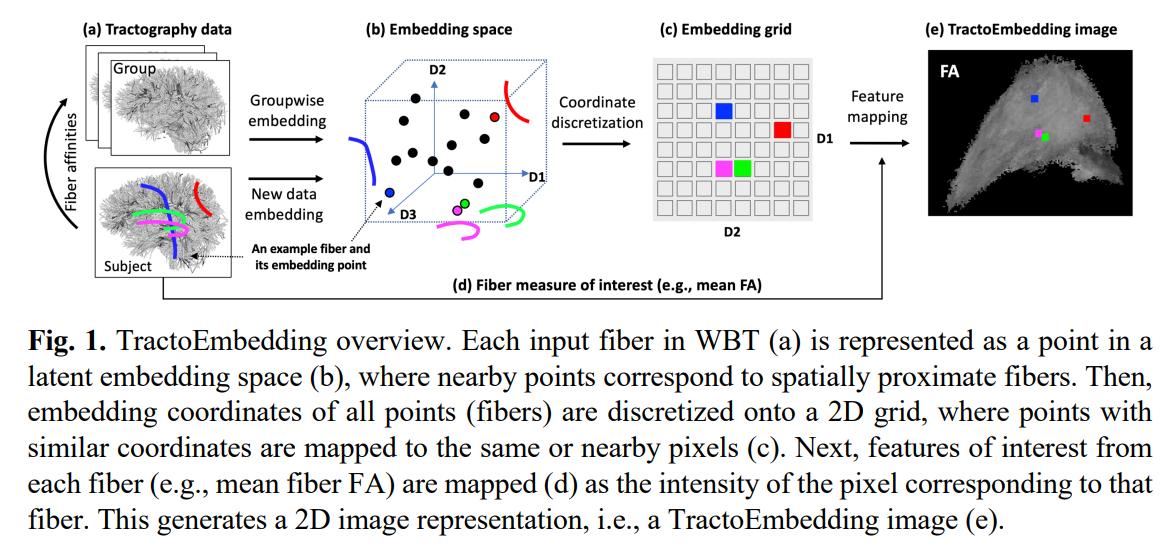
TractoEmbedding
-
Spectral embedding is used to create the latent space, using random samples of fibers from the HCP dataset. Then, to embed new fibers to the latent space, the fibers are registered to the same space as the rest and then embedded using spectral embedding still.
-
The coordinates of each embedded fibers are discretized to a 2D grid (taking only the first two dimensions of the latent space). The size of the 2D grid defines the resolution of the image in the third step.
-
Each point in the 2D grid is colored according to a metric computed on its corresponding fiber (mean FA, for example). If more than one fiber corresponds to the same pixel, the values are averaged. Multiple images for different fiber metrics can be computed, at different resolutions. Multiple images can be produced for left and right hemisphere and commissural fibers.
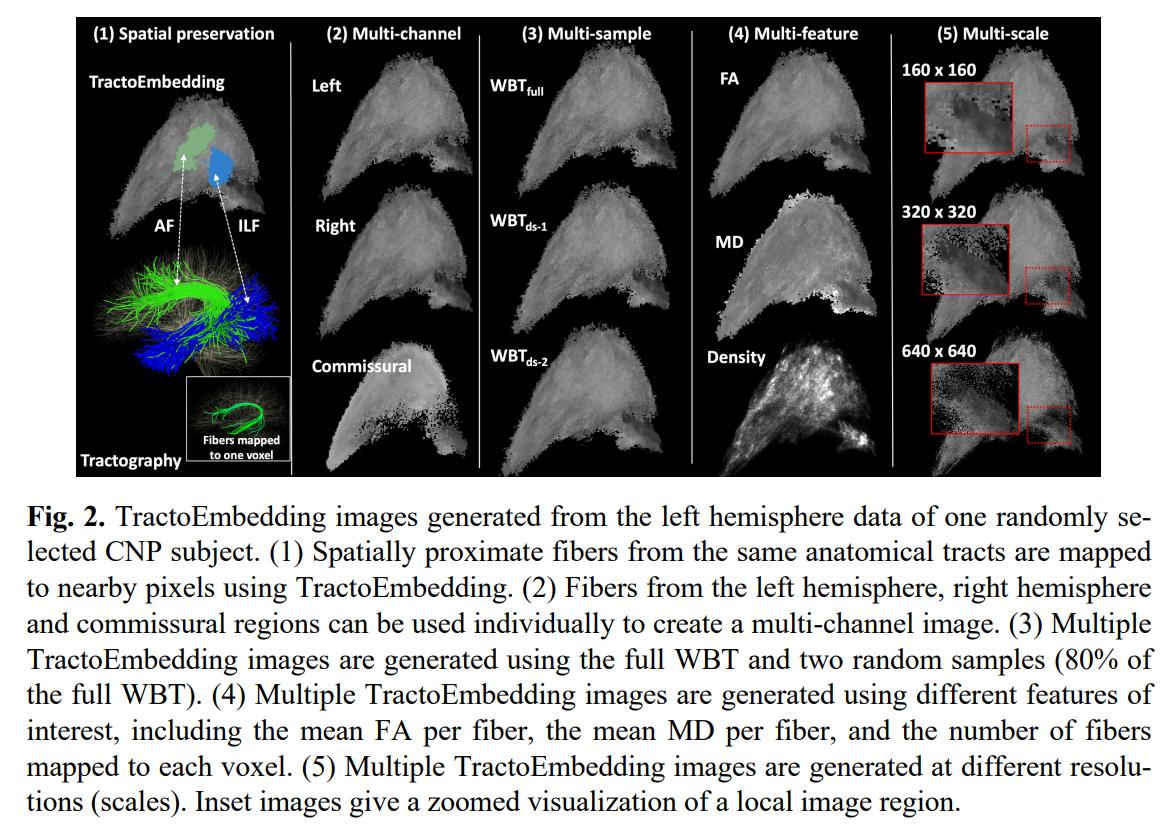
The authors argue TractoEmbedding has multiple advantages:
- Produces a 2D image that preserves the spatial relationship of fibers that can be leveraged by CCNNs or ViTs
- Multi-channel representation where each channel corresponds to a brain region
- Multiple embeddings can be generated by selecting random samples of fibers
- Can be used to encode any fiber metric
- Allows for fiber representations at different scales
TractoFormer
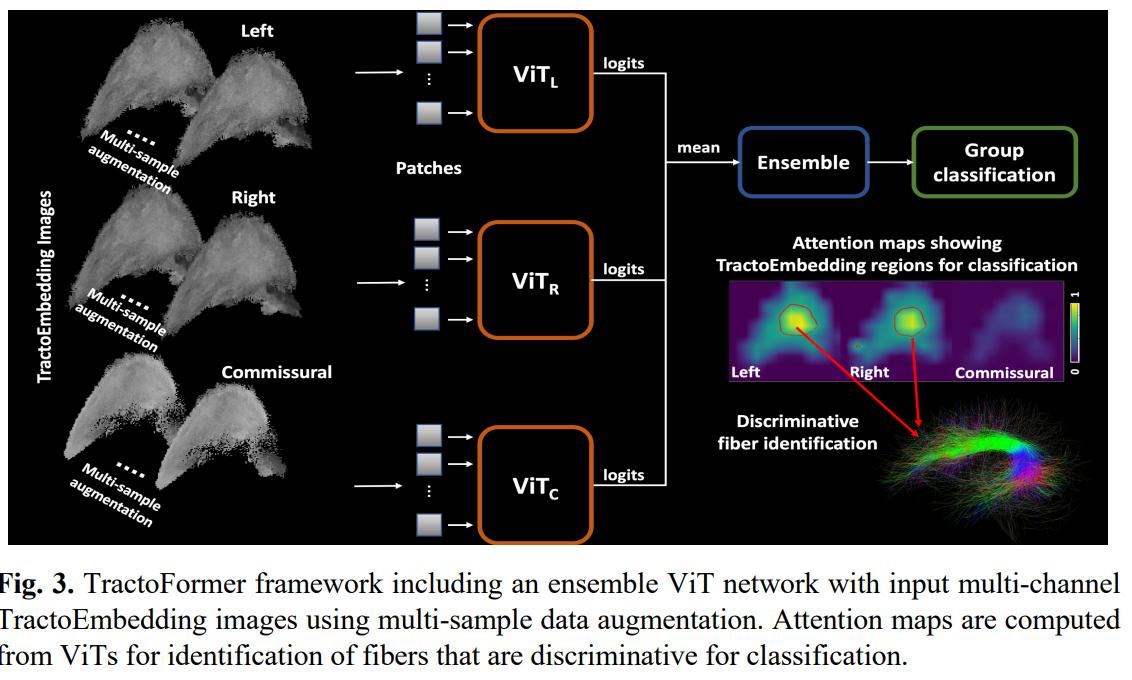
An ensemble of 3 ViTs (one for each type of fiber) is used for classiication. Multiple samples of fibers are used to generate images, to alleviate the hungriness of ViTs for data.
Tasks
Two tasks are considered to test the discriminative powers of TractoFormer using TractoEmbedding.
-
The authors add gaussian noise to the mean FA of fibers of two copies (G1 and G2) of the 103 healthy subjects of the second dataset. Then, the mean FA of one bundle (the cortico-spinal tracts, CST) of G2 is altered. This first task is to test if TractoFormer can discriminate between groups, and which fibers contribute to the differences.
-
Classification between healthy and schizophrenic subjects using the second dataset.
Results

TractoFormer was able to achieve 100% accuracy in the first task. Moreover, the attention maps succesfully identified CST tracts. For the second task, results are a bit less conclusive:
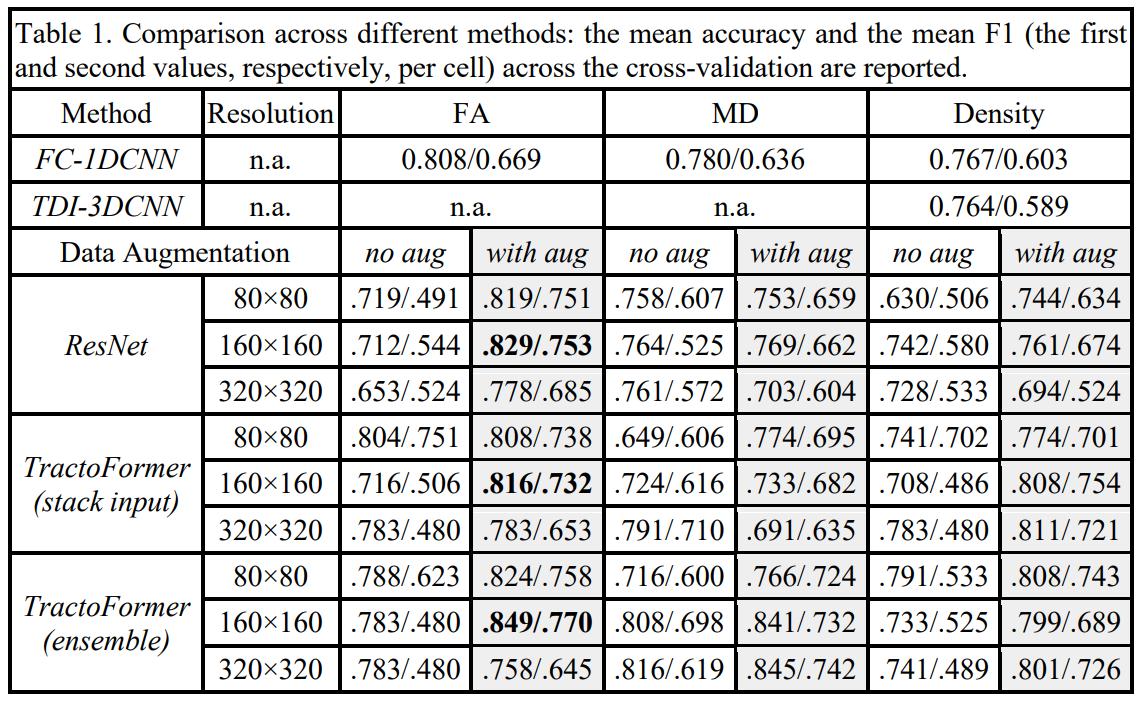
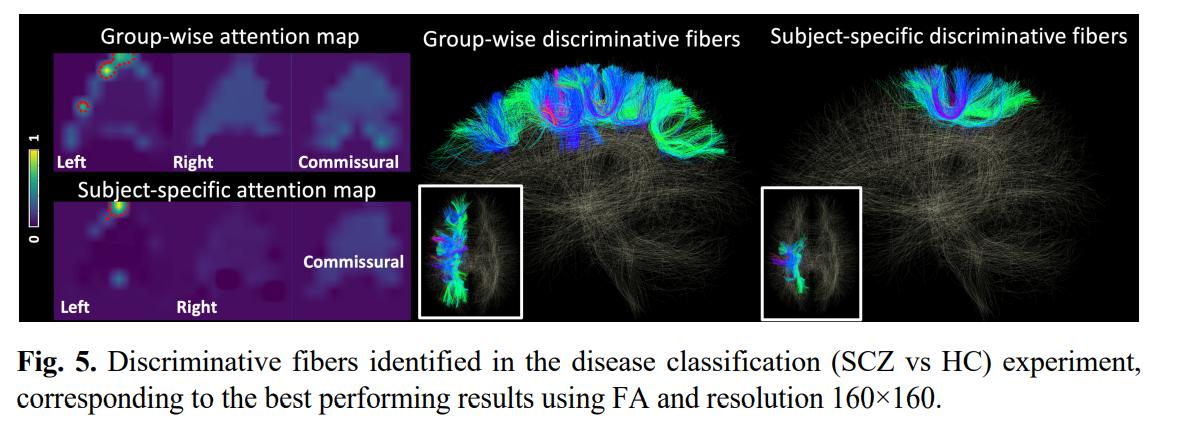
Multiple studies have suggested these white matter regions are affected in SCZ
Conclusions
TractoFormer can use information from single fibers to do classification and its attention mechanism helps interpretability. TractoEmbedding allows the use of “vision” architectures for this kind of tasks.