Neuro4Neuro: A neural network approach for neural tract segmentation using large-scale population-based diffusion imaging
Highlights
Authors present a 3D U-Net-based deep learning method for bundle segmentation using diffusion information.
Introduction
WM fibers reconstructed by tractography can be automatically grouped into bundles that match the known functional and/or structural role of these groups. Different strategies exist for automatic bundle segmentation. According to the authors these can be classified into semi-automatic, atlas-based and clustering methods.
Convolutional Neural Networks (CNN) have emerged as a powerful tool and shown superior performance for segmentation tasks across different domains.
Methods
Authors propose a 3D U-Net model with PReLU non-linearities.
They performed a number of experiments to find the combinations of input data that provided the best results. The best results were obtained using the diffusion tensor coefficients as the input data.
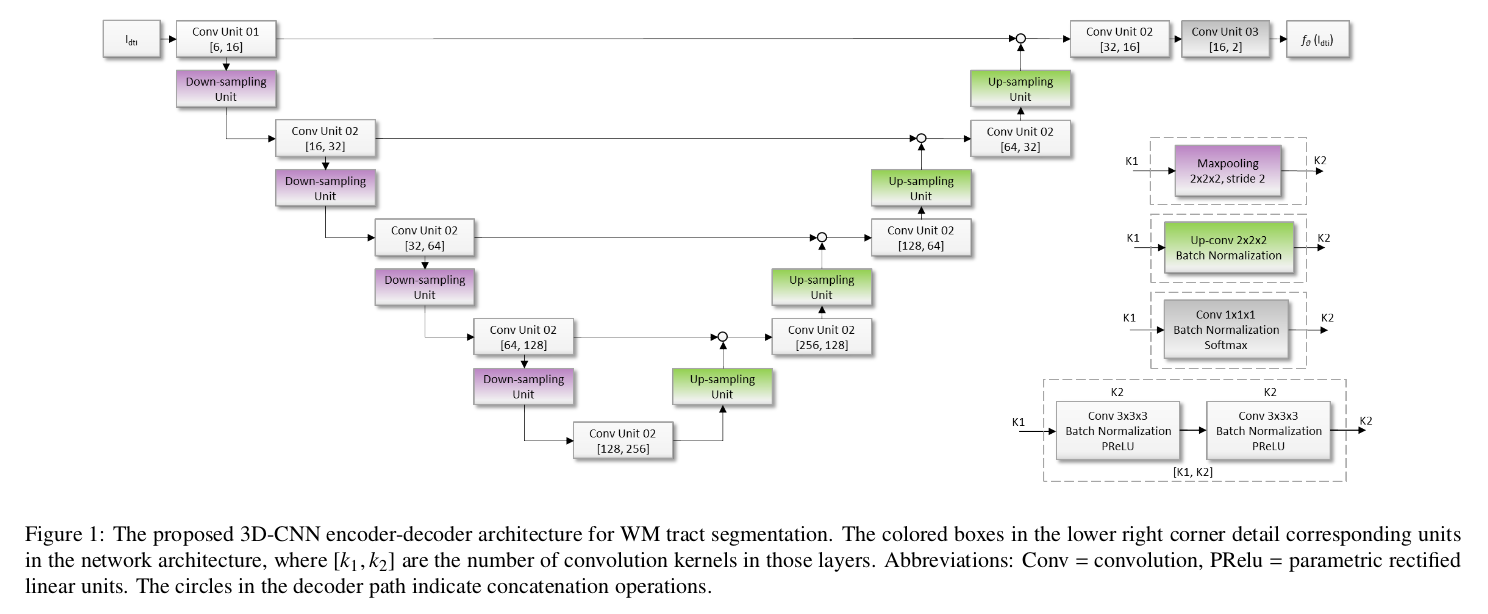
The reference for training and evaluation was based on an atlas-based probabilistic tractography pipeline. The resulting tract-specific density images were normalized by division with the total number of tracts in the tractography run. Tract-specific thresholds were established by maximizing the FA reproducibility on a training set of 30 subjects with 2. Outliers scans were discarded after visual inspection/quality control.
For each bundle, a ROI was defined based on the reference segmentation.
A separate model was trained for each bundle generating a binary output.
The loss is the weighed inner product, which removes the log operation wrt the cross-entropy. The weight allows to trade between precision and recall.
The performance measure is the Dice score measured within the ROI of each bundle.
Data
The Rotterdam Study (focuses on causes and consequences of age-related diseases) was used. 9752 dMRI scans featuring 25 gradient encoding directions at \(b = 1000 s/mm^2\) were used.
25 bundles were considered.
Results
The average segmentation accuracy was DC = 0.74 over 25 tracts, with the best performance in the medial lemniscus tract (DC = 0.84).
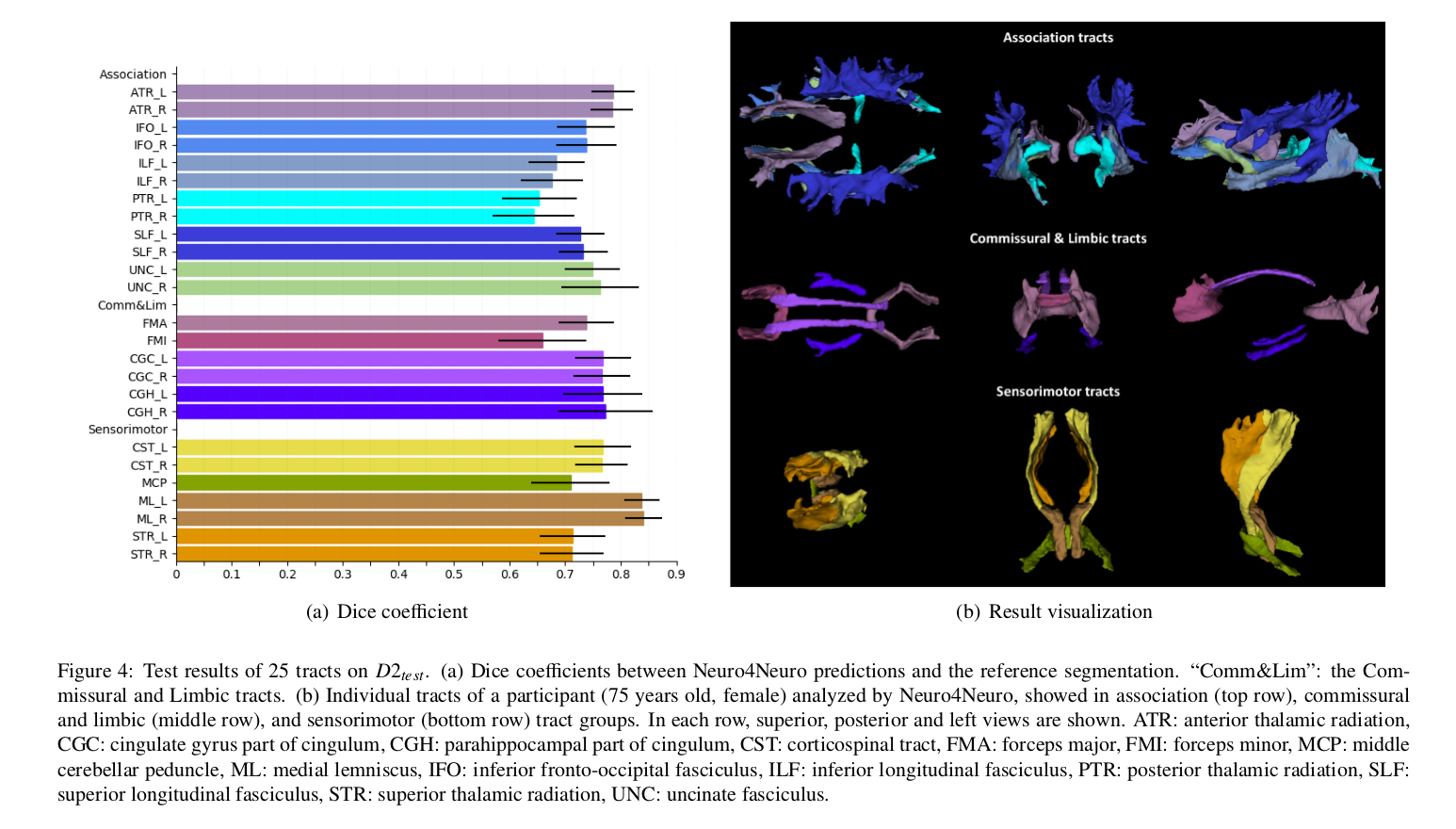
Authors also conducted a test-retest reproducibility study.
The regression coefficients between DTI measures (FA, MD) and the proposed and the reference method were also computed.
An additional dataset from the Iris Study was used to test the generalization ability of the method. Results of the bundle segmentation for healthy and different dementia groups were included.
Conclusions
The main advantages of the proposed method are:
- Acceleration wrt to conventional methods.
- Avoiding additional steps required by other methods, such as parcellation, atlas registration and fiber tracking.
Comments
- The choice of the input providing the best results and other hyper-parameters were set based on experiments done on a single bundle, the forceps minor. As acknowledged by the authors, this is a limitation (or a compromise taken for practical reasons).
- The literature review includes some works that did not deal with tract segmentation.
- The advantage over Wasserthal’s TractSeg method is unclear (besides not requiring three different networks for each anatomical axis, i.e. being 3D over 2D), and not discussed.
- A separate U-Net model is required to segment each bundle.
- The Dice score scale in Figure 4 does not cover the full [0, 1] range, which may give an overly optimistic performance impression unless properly read.