Estimating localized complexity of white-matter wiring with GANs
Introduction
Authors propose a measure that considers the complexity of a voxel in its local context (region) to quantify the localized wiring complexity of the white matter. According to them, this would allow, for example, to identify “particularly ambiguous regions of the brain for tractographic approaches”.
Authors define the localized wiring complexity of white matter as how accurately and confidently a well-trained model can predict the missing patch.
Authors seem to translate the wiring complexity as “uncertainty along major neuronal pathway” in a given diffusion Magnetic Resonance Imaging (dMRI) feature, and call that a “missing patch”.
Methods
Authors try to regress the multidirectional anisotropy (MDA) feature vector in the “missing patches” of their input dMRI images.
Authors use a conditional GAN to accomplish the task. Their discriminator loss is “typical” (thus assumed to be the binary cross entropy); the loss used for the generator is the one shown in Eq. 1:
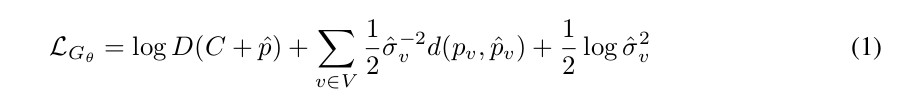
where \(v\) indexes the set of voxels in the considered \(V\) patch; \(d\) is an \(L2\) norm distance between the input patch and the predicted patch; and \(D\) is the probability that the discriminator outputs for the generated patch.
Data
630 subject data from the Human Connectome Project (HCP) dataset: 442 for training; 94 for validation; and 94 for testing.
Results
Figure 1 shows the location of the patch of interest, the ground-truth and the model output.
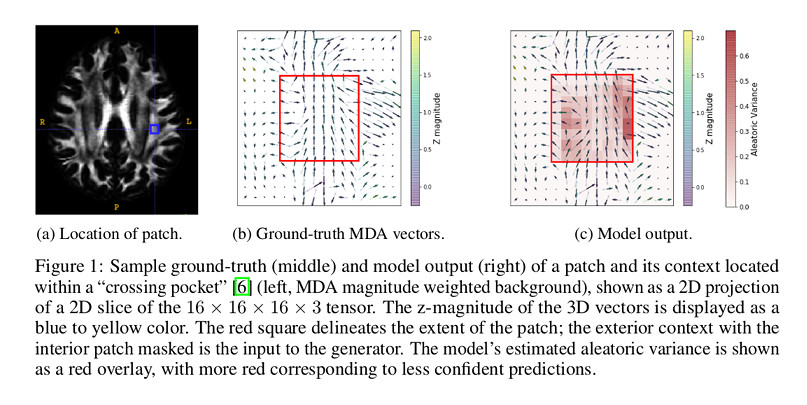
Figure 2 shows the variance predicted by the model.
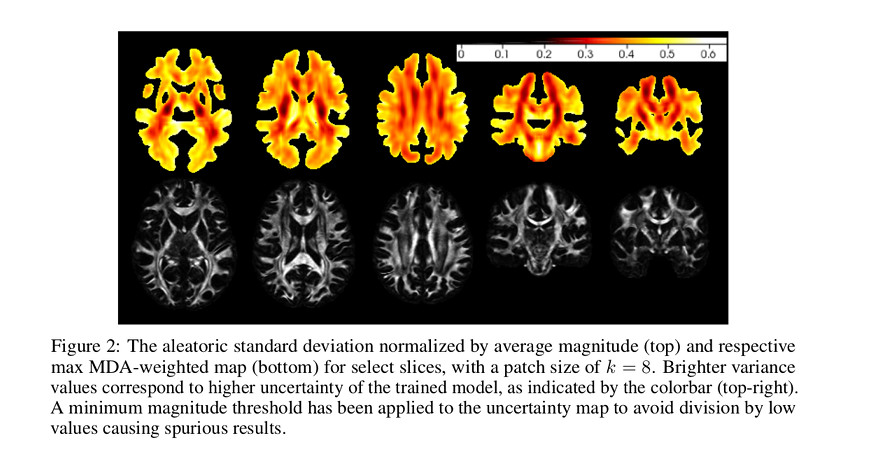
According to the authors “uncertainty is lowest within major fiber bundles, which are relatively simple uniform structures, though greater near junctions or crossings of those bundles”.
Conclusions
Authors mention that their future work includes to “aim to better quantify and understand heterogeneity, especially in the presence of complex wiring patterns.”
Comments
- It is unclear the relationship of their problem to image inpainting methods that authors mention.
- It is unclear what the input to their network is.
- It is unclear how they built their ground-truth.
- MDA is not a feature that is used commonly in dMRI analysis, at least not under that denomination.
- The paper lacks a better rationale and explanation about their model/network architecture.
- No quantitative results are presented.
- No comparisons against other methods are presented to allow users to assess the performance of their method.
- The message of the paper is unclear.
- The study uses healthy subject data; a higher “wiring complexity” is usually assumed to be present in lesion brain populations, so such populations would may be fit better for this kind of analysis.