q-Space Novelty Detection with Variational Autoencoders
Highlights
Authors propose an Variational Autoencoder (VAE) for novelty detection (detection of cases that have not been seen during training) in dMRI scans of multiple-sclerosis (MS) patients.
Their results outperforms their baseline when identifying lesion voxels.
Introduction
The purpose of novelty detection is to score how dissimilar each test sample is from a “normal” training set. Since abnormal samples are not used during training, novelty metrics are defined based on the assumptions that
- The VAE is less capable of reconstructing abnormal samples well,
- Abnormal samples more strongly violate the VAE regularizer,
- Abnormal samples differ from normal samples in the input-feature space, the VAE latent space and VAE output.
Thus, authors propose to use novelty scores to detect dMRI scans where disease is present. The distance between the test data points and its nearest neighbor from the reference dataset is used as the novelty score.
Methods
Authors propose a variety of novelty scores based on different criteria, such as distances between data points or distributions involved in the VAE formulation, the distance in different spaces, etc.
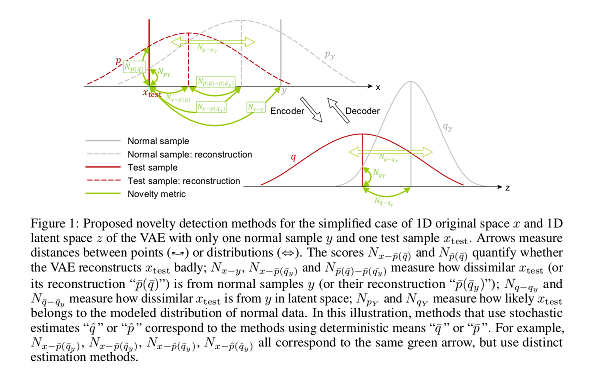
A summary of the scores they propose:
- Novelty in latent space
- VAE regularizer score
- Latent-space distances
- Euclidean distance between means of distributions
- Bhattacharyaa distance between distributions
- Density-based distance in latent space
- Novelty in original feature space
- VAE reconstruction-based
- Deterministic reconstruction error
- Deterministic reconstruction likelihood
- Encoder-stochastic reconstruction error
- Encoder-stochastic reconstruction likelihood
- Decoder-stochastic reconstruction error
- Fully-stochastic reconstruction error
- Distance- and density-based
- Distance to the closest (VAE decoder-) generated sample (from any latent vector)
- Additive inverse of highest generated likelihood
- VAE reconstruction-based
- Novelty as VAE loss
Their baseline is another novelty detection method not based in deep learning by the same authors.
Data
- 26 dMRI acquisitions of healthy volunteers for training: 46 volumes (6 B0, 40 diffusion directions); 3 dMRI acquisitions of MS patients (same acquisition parameters) + manually annotated lesions for testing.
- MNIST: single digit as novel; training on remaining ones.
Results
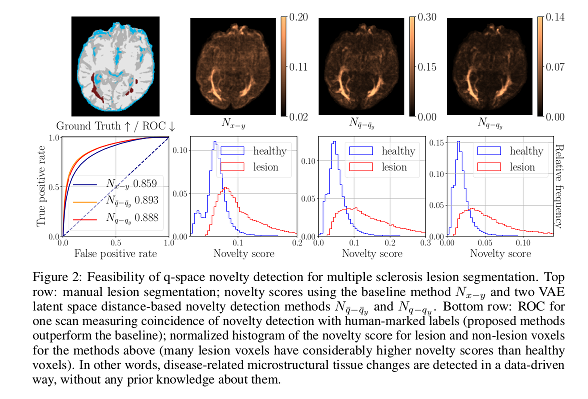
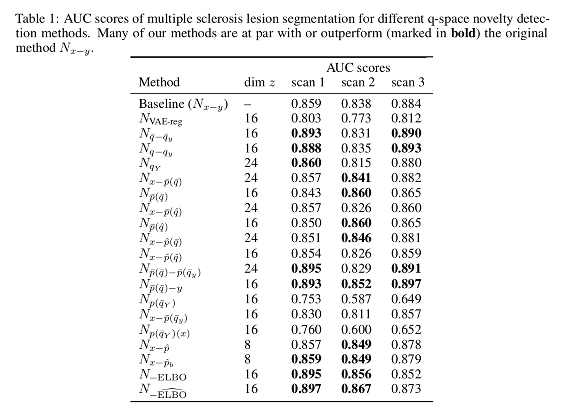
For the dMRI dataset, the proposed methods show a “good performance” when compared to their baseline. Novelty scores based on Euclidean distances in the latent space outperform those based in distances in the original space.
Reconstruction-based methods perform well provided that the dimensionality of the latent space has enough capacity.
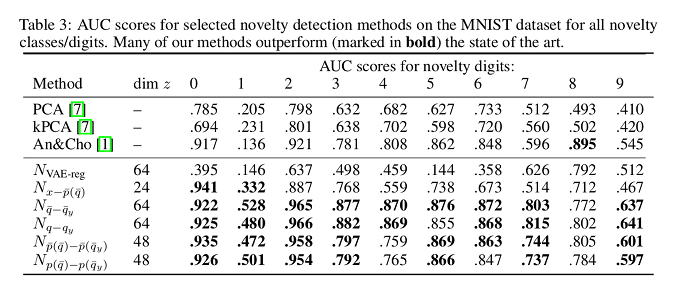
For the MNIST dataset, results show greater variance. Scores for digit “1” are considerably worse than for the rest, probably because the novelty presented by that digit is implied in other digits used for training.
Conclusions
A set of methods were presented for novelty detection in MS dMRI scans.
Remarks
No comparison is shown against supervised methods.
Any conclusion on the dMRI dataset seems to be limited due to the limited number of scans used for testing. Furthermore, according to the novelty score graphs shown, the distinction of the healthy and lesion tissue does not seem to be conclusive.
In order to compare the scores, rather than reporting results per each scan, reporting statistical performances (e.g. meand an variance) would be more meaningful.
The discussion lacks sound arguments as to why a novelty score is “better” than another one beyond statements that seem to be general for a VAE.