Missing MRI Pulse Sequence Synthesis using Multi-Modal Generative Adversarial Network
Highlights
- Authors propose to use a Generative Adversarial Network (GAN) to generate MRI pulse sequences that have not been acquired during a study.
- The comparison to unimodal and multi-modal methods shows that their method outperforms both quantitatively and qualitatively.
Introduction
Different MRI contrasts provide different anatomical information. This contrast may be achieved by varying the gradient pulse sequence echo time (TE) and repetition time (TR). The most common contrasts are T1-weighted (T1), T2-weighted (T2), contrast-enhanced T1 (T1c or CE T1), diffusion weighed (DW) contrast, and T2 Fluid-attenuated Inversion Recovery (FLAIR).
Due to various constraints (time, patient conditions, etc.) it may not be possible to acquire all sequences.
Synthesis methods are classified as:
- Unimodal: both the input and output of the system is a single pulse sequence. Usually involves using atlases for intensity mapping.
- Multimodal: multiple input sequences are used to synthesize a single or multiple sequences.
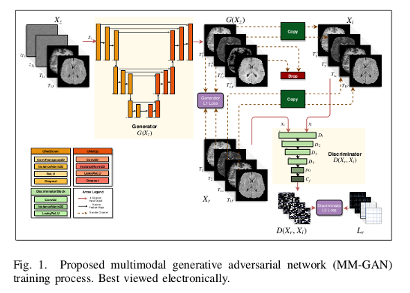
A Multi-Modal Generative Adversarial Network (MM-GAN) capable of synthesizing “missing” sequences in a single forward pass through the network by combining information from all available sequences is proposed.
Methods
The presented method differs from other approaches in the sense that their output is truly multi-modal (many sequences can be reconstructed), and that the missing sequence can vary across experiments.
Their implicit conditioning to produce a particular type of sequences is achieved through:
- Noise imputation: the input to the generator contains noise at the place of missing sequences.
- Sequence-selective loss computation in the generator: the reconstruction loss is computed only between the synthesized sequences for the generator and ignores the sequences that are present.
- Sequence-selective discrimination: the adversarial loss is penalized only for incorrectly synthesizing the missing sequences, and not for incorrectly synthesizing the sequences that were already present.
- Curriculum learning: the difficulty of examples increases as more sequences go missing.
The input to the network is a 4-channel (corresponding to 4 sequences) 2D axial slice, where random Gaussian noise is imputed for channels corresponding to missing sequences. The output of the network is a 4-channel 2D axial slice, in which the originally missing sequences are synthesized by the network.
A variant of Pix2Pix architecture1 is used. The generator is a UNet and the discriminator has a PatchGAN architecture2.
Data
- Ischemic Stroke Lesion Segmentation Challenge 2015 (ISLES2015)
- Multimodal Brain Tumor Segmentation Challenge 2018 (BraTS2018)
Results
Baselines:
Main results:
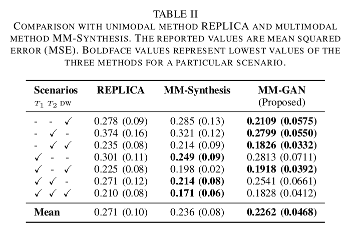
- The proposed MM-GAN model performs consistently better than the baseline when synthesizing just one sequence.
- Using CL based learning did not help in multi-input/single output experiments. They state that it is due to the presence of some “highly informative” sequence that dominates the synthesis.
- However, it did help achieving a stable training of the network in the multi-input/multi-output setting.
- As more sequences start missing, the task of synthesizing missing sequences gets harder.
- The proposed method performs consistently better than REPLICA, and gets a lower overall score than MM-Synthesis.
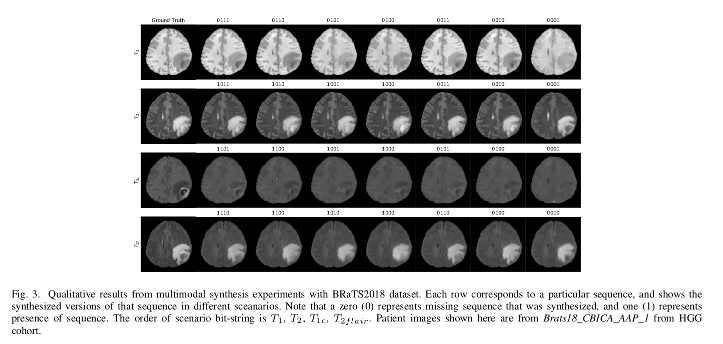
- The synthesis of the tumor in the final images depend heavily on the available sequences.
Conclusions
The work presents a truly multi-modal method that is multi-input and multi-output, generalizing to any combination of available and missing sequences.
The method outperforms the multimodal synthesis methods used as baselines.
References
-
Guha Balakrishnan, Amy Zhao, Mert R. Sabuncu, John Guttag and Adrian V. Dalca. An unsupervised learning model for deformable medical image registration. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 9252-9260, 2018 ↩
-
Phillip Isola, Jun-Yan Zhu, Tinghui Zhou, and Alexei A. Efros. Image-to-image translation with conditional adversarial networks. The IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 1125-1134, 2017. ↩
-
Salman Ul Hassan Dar et al. Image Synthesis in Multi-Contrast MRI with Conditional Generative Adversarial Networks. IEEE Transactions on Medical Imaging. pp. 1-1, 2019. ↩
-
Amod Jog, Aaron Carass, Snehashis Roy, Dzung L. Pham, and Jerry L. Prince. Random forest regression for magnetic resonance image synthesis. Medical Image Analysis, vol. 35, pp. 475-488, jan 2017. ↩
-
Agisilaos Chartsias, Thomas Joyce, Mario Valerio Giuffrida, and Sotirios A. Tsaftaris. Multimodal MR Synthesis via Modality-Invariant Latent Representation. IEEE Transactions on Medical Imaging, vol. 37, no. 3, pp. 803-814, mar 2018. ↩