3D Whole Brain Segmentation using Spatially Localized Atlas Network Tiles
Highlights
- Authors propose a 3D patch-based multiple FCN method for brain parcellation.
- The work presented uses a multi-atlas approach to meet the goal.
Introduction
Multi-atlas and path-based segmentation methods overcome the single-atlas performance issues derived from inter-subject anatomy variability.
2D-based CNN yield inferior spatial consistency on the third dimension.
Fitting whole brain high resolution MRI scans in memory is impractical.
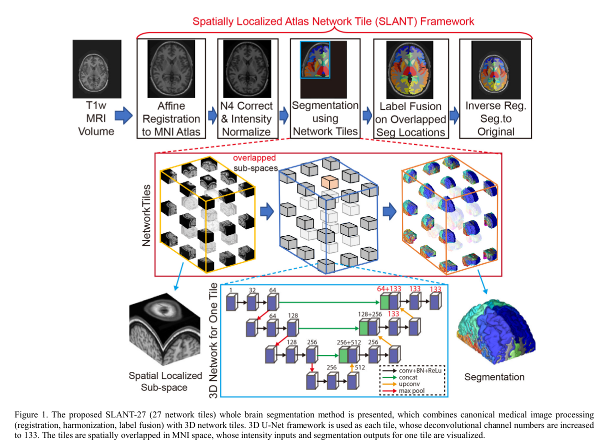
Authors propose a Spatially Localized Atlas Network (SLANT) that uses multiple, independent 3D FCN each of which processes only a region of the volume.
Methods
Brains are registered to a common space and intensities are normalized.
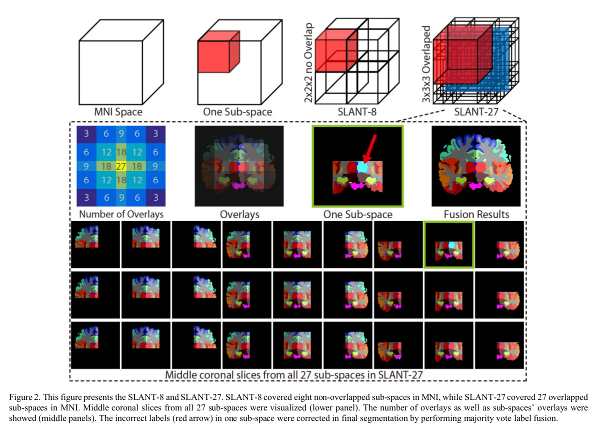
The brain is divided into tiles. Two variants are presented:
- 8 non-overlapping sub-spaces.
- 27 overlapping sub-spaces.
The same 3D U-Net architecture is used for each tile.
A majority vote fusion is used to get the final segmentation in the overlapping case.
Training of each network tile is done on 5111 initially unlabeled scans using an initial set of 45 atlases. A smaller set of the 45 atlases is used for further fine-tuning each tiles.
Label fusion of atlases for the 5111 training scans is done using a non-local spatial staple label fusion (NLSS) by the same authors.
In the testing phase, a PCA-based atlas selection is used to fuse the labels of the training data together: a test scan is segmented using the 15 atlases with the least distance with respect to the scan in the projected PCA space.
Data
T1 MRI datasets come from 11 different studies.
5 scans from the OASIS dataset for validation; 14 datasets (1 from Colin27 and 13 from CANDI) for testing.
Benchmarking
Baselines:
- Joint Label Fusion (JLF)
- Non-local Spatial Staple (NNLS)
- Naive 3D U-Net
- HC-Net
Dice similarity coefficient and mean surface distance.
Results
27-day training on a NVIDIA TITAN GPU for 5111 scans.
Testing is relatively fast (~15 minutes for a single volume).
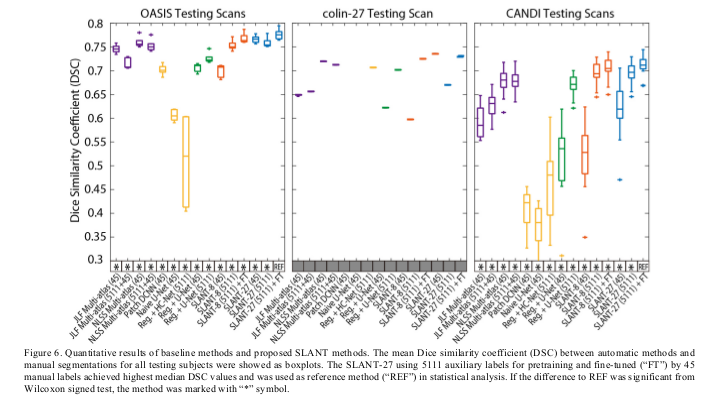
The SLANT-27 using 5111 auxiliary labels for pre-training and fine-tuned by 45 manual labels achieved highest median DSC value.
Conclusions
The computational time increases linearly with the number of tiles when a single GPU is used.
Remarks
The reason behind using a different fusion strategy for the additional 5111 training scans (the NLSS method) and the testing scans (a PCA-based strategy) is unclear.
The number of testing images is rather low.
The method requires a non-negligible amount of computational resources.
The choice of the patch size was empirically found to provide improved results over other choices (e.g. SLANT-64).