Data augmentation using learned transforms for one-shot medical image segmentation
Highlights
- Authors propose an automated data augmentation method for medical images focusing on the one-shot segmentation scenario for Magnetic Resonance Imaging (MRI) brain scans.
- The model of transforms is learned from the images, and used along with the labeled example to synthesize additional labeled training examples for supervised segmentation.
- Rather than removing intensity variations, it enables a segmentation method to be robust to the natural variations in MRI scans.
Introduction
Labeling datasets of medical images requires significant expertise and time, and is infeasible at large scales. These techniques involve costly engineering efforts, and are typically dataset-specific.
The problem of limited labeled data is exacerbated by differences in image acquisition procedures across machines and institutions, which can produce wide variations in resolution, image noise, and tissue appearance.
When large datasets are required in deep learning training contexts, pre-processing steps and data augmentation are usually required.
Data augmentation functions, such as random image rotations or random non-linear deformations are easy to implement, and have been shown to be effective at improving segmentation accuracy in some settings. However, these functions have limited ability to emulate diverse and realistic exam image features.
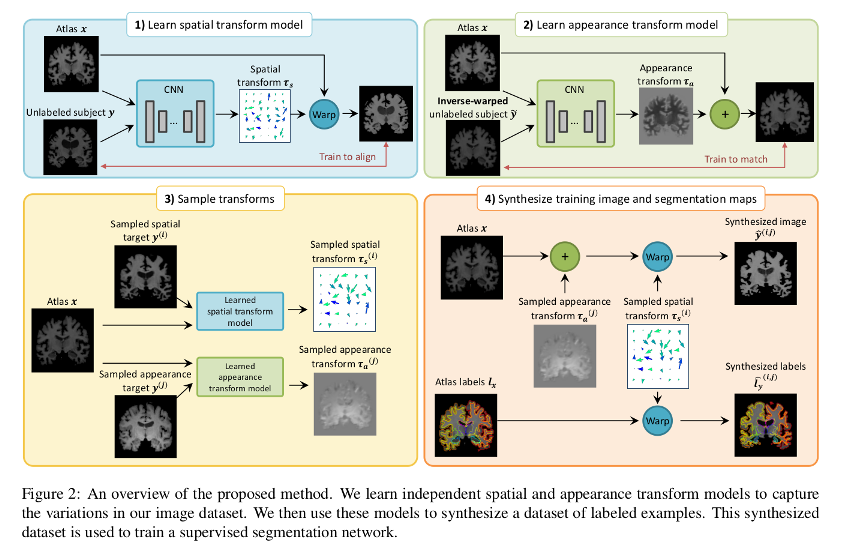
Authors propose to address the challenges of limited labeled data by learning to synthesize diverse and realistic labeled examples, using learning-based spatial and appearance transforms between images in the dataset.
New examples are synthesized by sampling transforms and applying them to a single labeled example. The method is tested on the task of one-shot segmentation of brain MRI scans.
Methods
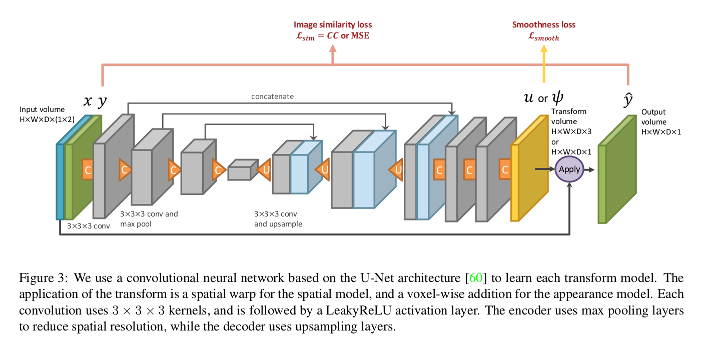
Each transform is comprised of a spatial deformation field and an intensity change. The transforms are learned by separate networks to capture the distribution of anatomical and appearance differences between the labeled atlas and each unlabeled volume.
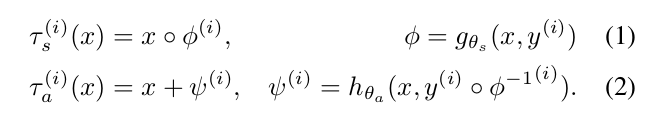
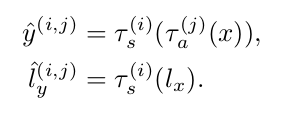
The transformation functions’ parameters are optimized separately using CCNs:
- The spatial transformation network used is a variant of their VoxelMorph1 with normalized cross-correlation as the image similarity loss.
- The appearance loss is a Mean Squared Error (MSE) that incorporates a smoothing regularization term.
Labeled synthetic volumes are generated by applying the transforms computed from the target volumes to the labeled atlas. These new labeled training examples are then included in the labeled training set for a supervised segmentation network.
Data
T1-weighted MRI brain scans from the dataset described in Balakrishnan et. al1.
Segmentation maps are obtained using FreeSurfer2.
101 scans are randomly selected to be present at training time, and a single labeled volume is used among these as the label set for the training. 50 additional scans are used as the validation set, and 100 others are used as the test set.
Results
The method is compared against four baselines:
- Single-atlas segmentation (SAS)
- Data augmentation using single-atlas segmentation (SAS-aug)
- Hand-tuned random data augmentation (rand-aug)
- Fully supervised segmentation
Consistent improvement of Dice scores across all brain structures are reported.
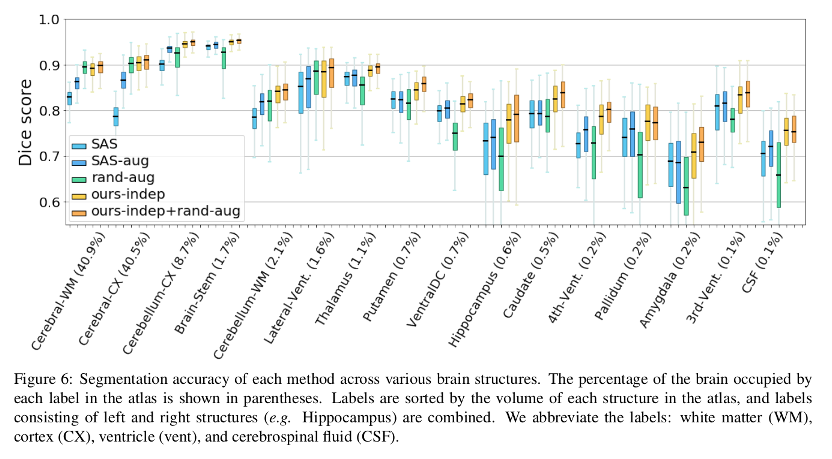
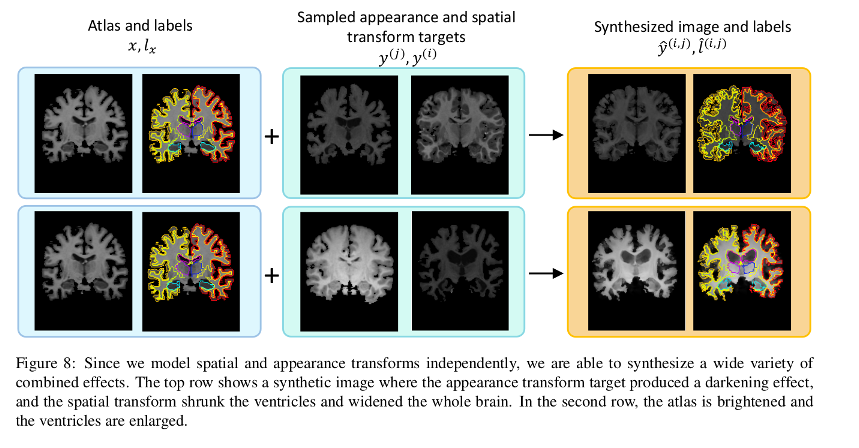
Conclusions
Authors demonstrate a method for data augmentation generating new volumes by sampling spatial and appearance transforms and applying them to the labeled volume. The synthesized examples are used to train a supervised segmentation model that shows an improved performance over the compared methods.
Remarks
- Even if a single labeled instance is used at the beginning, labeled sets are augmented, so it is not a real single-shot method.
- No comparison to other deep learning-specific synthetic data generation (e.g. GANs) methods is provided.
- The dataset and/or division used to train the transformation learning networks is unclear.
References
-
Guha Balakrishnan, Amy Zhao, Mert R. Sabuncu, John Guttag and Adrian V. Dalca. An unsupervised learning model for deformable medical image registration. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 9252-9260, 2018 ↩ ↩2
-
FreeSurfer: https://surfer.nmr.mgh.harvard.edu/ ↩