Spherical Harmonic Residual Network for Diffusion Signal Harmonization
Summary
Goal: Harmonize the dMRI signal differences between scanners
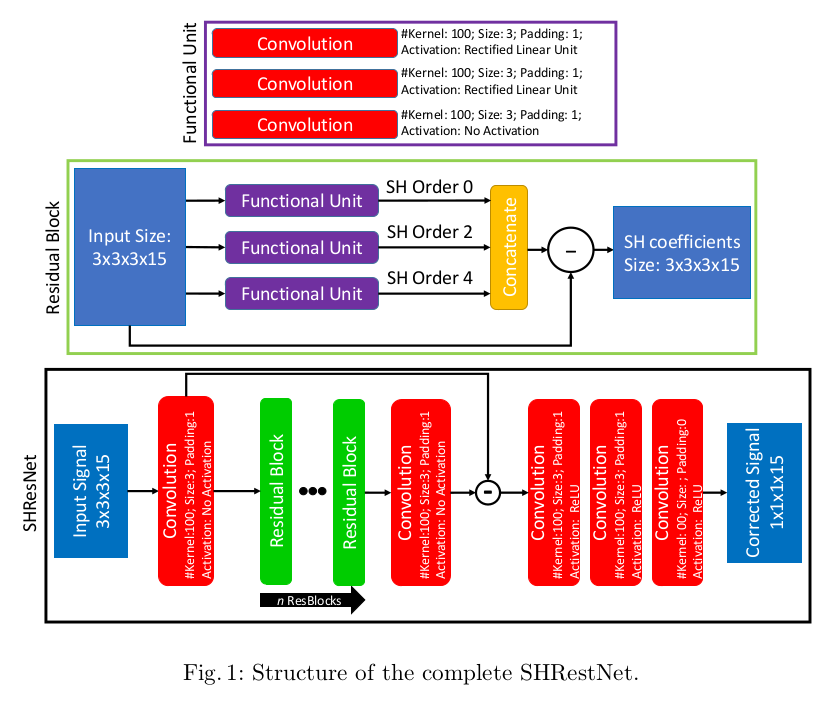
Method: Use a ResNet on 3x3x3 patches to harmonize SH coefficients voxel-wise, by learning a mapping between sites.
Highlights:
- No need for registration (at test time)
- Model-free (spherical harmonics do not impose a model on the data)
Model
- RISH projection: At test time, RISH features are used to project the harmonized signal and correct for changes in fiber orientation
where \(S_i'\) are the hamonized SH coefficients and \(S_i\) the non-harmonized SH coefficients of order \(i\).
Reminder:
RISH: Squared L2 norm for each order of SH coefficients
Results
Datasets:
CDMRI harmonization Challenge
- 10 healthy subjects, scanned on a 3T GE Excite-HD and a 3T Siemens Prisma, with 30 directions at b=1200.
Training:
- FAST registration for training subjects
- 5 epochs using Adam with batch size 256, then SGD with batch size 128 and learning rate decay…
Experiments:
- Investigate the sensitivity of the method to the number of ResBlocks n (Table 1)
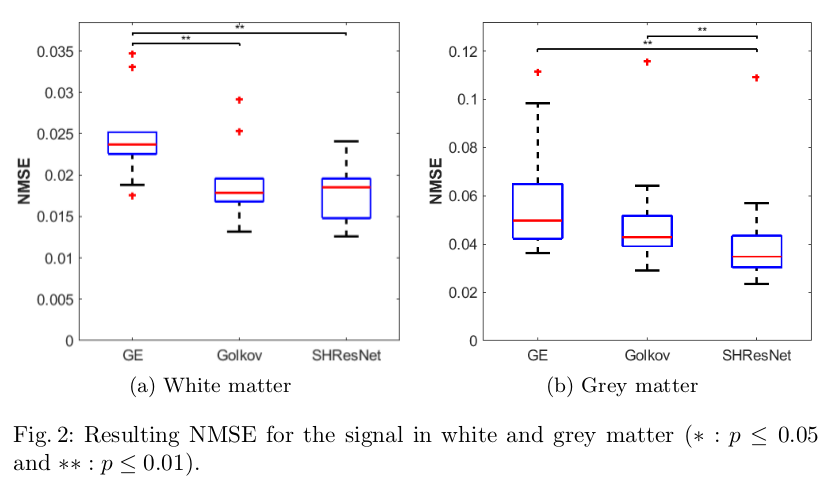
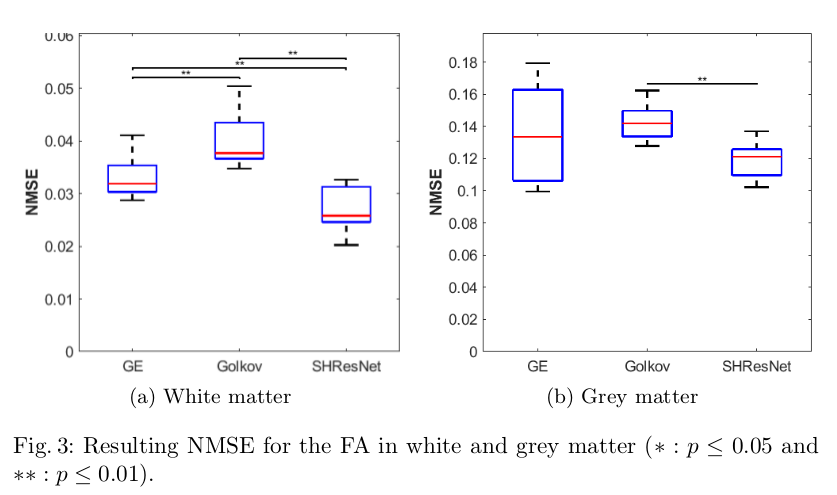
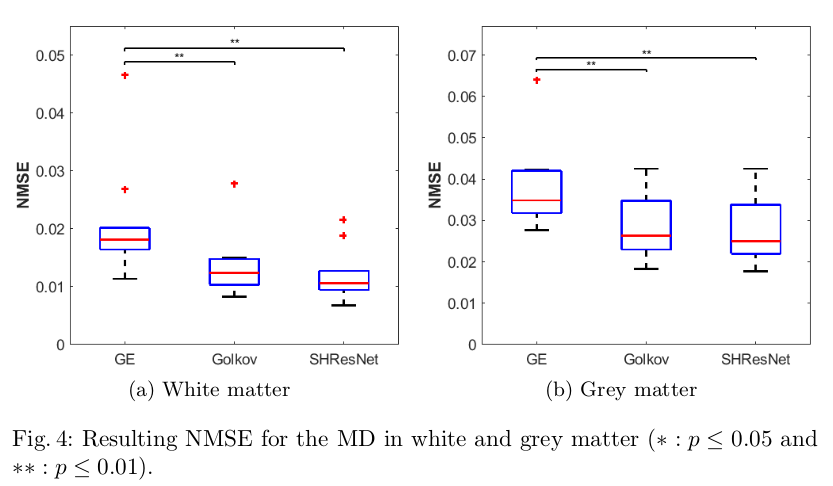
- Evaluate the ability to reduce inter-scanner variance in the signal itself and metrics like FA/MD (Figures 2,3,4)
- 10-fold cross-validation using the 10 subjects (8 training, 1 validation, 1 test).
The only reported baseline is the Golkov method1, a 3-layer, 150 units neural network with ReLU and Dropout designed, applied on a single voxel at a time; it was “designed” for the estimation of scalar measures from diffusion data, and seems completely unrelated to harmonization.
There is no classical method baseline…
-
Golkov, V., Dosovitskiy, A., Sperl, J.I., Menzel, M.I., Czisch, M., Sämann, P., Brox, T. and Cremers, D., 2016. Q-space deep learning: twelve-fold shorter and model-free diffusion MRI scans. IEEE transactions on medical imaging, 35(5), pp.1344-1351. ↩