An (almost) instant brain atlas segmentation for large-scale studies
Highlights
Authors propose a convolutional neural network (CNN) trained on the output produced by the state of the art models for whole brain segmentation on human brain MRI images.
Evaluation was done on healthy control and schizophrenia subject images.
Introduction
The network is based on a previous architecture from the same authors, called MeshNet1, which has been demostrated to perform successfully in gray and white matter segmentation.
They extend MeshNet to perform human brain atlas segmentation with 50 cortical, subcortical and cerebellar labels.
Methods
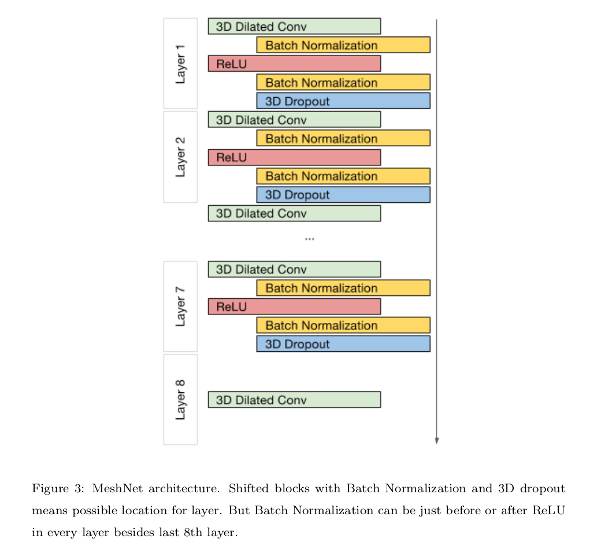
They propose an 8-layer CNN, and uses 3D convolutions to do the task.
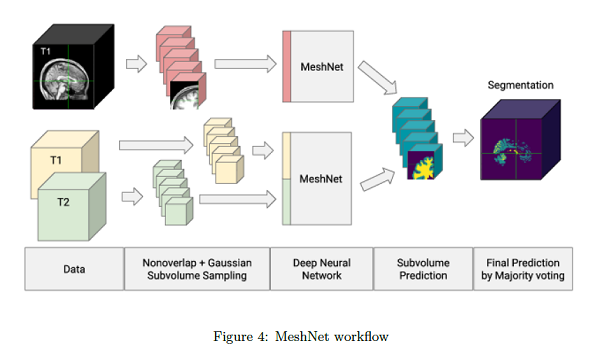
The use T1-weighed and T2-weighed data Human Connectome Project (HCP)2, and T1-weighed data from the multi-site Functional Bioinformatics Informatics Research (FBIRN)3 datasets.
Volume subsampling is required due to memory limitations. At training time, they divide the input volumes in non-overlapping Gaussian subsampled volumes and obtain a class prediction in every voxel. The final segmentation label for a tissue is obtained by voxel-wise majority voting.
The metrics they report on are:
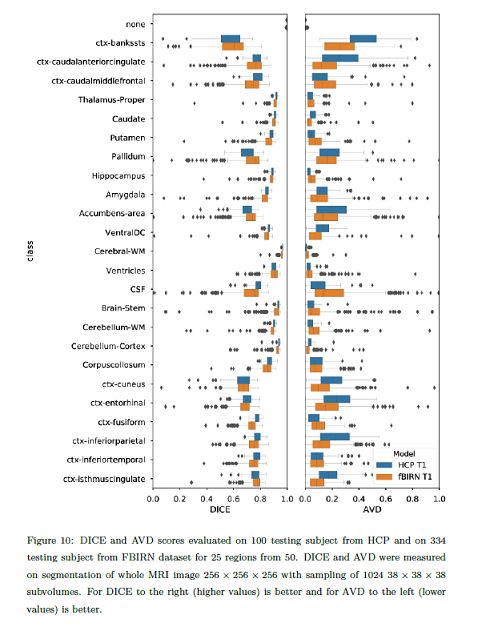
- The DICE coefficient for measuring spatial overlap.
- The Average Volume Difference (AVD) for validating volume values.
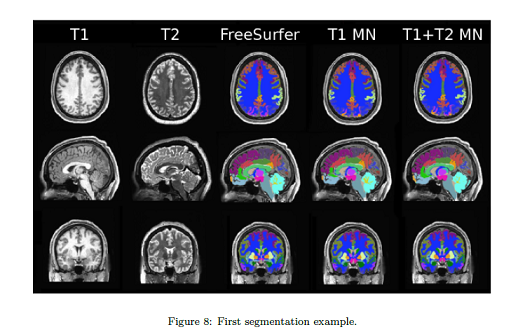
They benchmarking reference are the labels produced using Freesurfer4.
For the FBRIN dataset they used the HCP pre-trained weights, and hence they used a very reduced number of subjects for training and validation.
A statistical ANOVA analysis is also presented.
Results
The results are hard to generalize and vary a lot across brain regions.
They found among the network variants they tested, those that do not use dropout show a better overall performance.
Discussion
Authors claim to have proposed a method to perform very fast whole brain segmentations for large-scale studies using a fully convolutional deep learning network.
Comments
Their method requires knowing the voxels that belong to a structure.
The results are not clearly presented, and the conclusions are rather weak.
References
-
Alex Fedorov, Jeremy Johnson, Eswar Damaraju, Alexei Ozerin, Vince Calhoun, and Sergey Plis. End-to-end learning of brain tissue segmentation from imperfect labeling. In 2017 IEEE International Joint Conference on Neural Networks (IJCNN), 3785-3792, 2017. ↩