Deep Learning with Mixed Supervision for Brain Tumor Segmentation
Highlights
Authors propose a model to perform brain tumor segmentation and classification
jointly on Magnetic Resonance Images by training a deep learning network on
fully-annotated and weakly-annotated data.
The method is evaluated under the Multimodal Brain Tumor Segmentation BraTS 2018 Challenge framework.
Authors show that the proposed approach provides a significant improvement of segmentation performance compared to the standard supervised learning. Interestingly, the observed improvement is proportional to the ratio between weakly-annotated and fully-annotated images available for training.
Introduction
Convolutional Neural Network-based (CNN) methods have obtained the best performances on the four last editions of BraTS Challenge.
Most of the current state-of-the-art methods for tumor segmentation are based on machine learning models trained on manually segmented images.
Manually annotated data are time-consuming and costly. Furthermore, in medical imaging such tasks require high medical competences.
Images with a provided global label (indicating presence or absence of a tumor) are less informative but can be obtained at a substantially lower cost.
In the paper, authors propose to use a novel CNN-based segmentation model trained on fully-annotated (with provided ground truth segmentation) and weakly-annotated, with an image-level label indicating presence or absence of a tumor tissue within the image. They refer to this setting as “mixed supervision”. They propose to extend segmentation networks with an additional branch performing image-level classification.
The goal is to exploit the representation learning ability of CNNs to learn from weakly-annotated images while supervising the training using fully-annotated images in order to learn features relevant for the segmentation task.
Methods
Authors extend a segmentation CNN (U-net) with an additional subnetwork performing image-level classification and train the model for the two task jointly.
The U-net uses batch normalization and softmax activations.
The classification branch takes as input the second to last convolutional layer of U-net and is composed of one mean-pooling, one convolutional layer and 7 fully-connected layers. Mean pooling with ReLU activations are used before the last FC layers in the classification branch.
Binary and multi-class segmentation experiments are performed in the framework of the BraTS challenge. BraTS considers 4 classes (“non-tumor”, “contrast-enhancing core”, “edema”, and “non-enhancing core”).
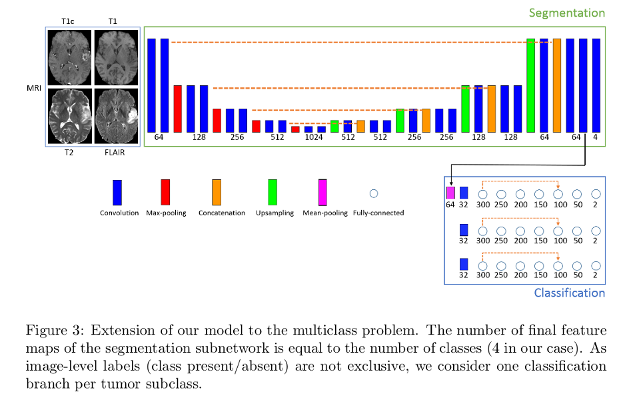
Binary segmentation
In order to train the model, weakly annotated images need to contain samples of datasets not containing tumors.
Training batches are composed of the three types of images available:
- \(k\) positive cases (containing a tumor) with provided segmentation.
- \(m\) negative cases (i.e. weakly annotated not containing tumor).
- \(n\) positive cases without provided segmentation (i.e. weakly annotated containing tumor).
Weighed pixel-level cross-entropy loss for segmentation task on \(k\) and $$m$. The weights are used to compensate class imbalance. They are set as the ratio between an empirical factor and the total number of pixels.
Pixel-level cross-entropy loss for classification task on all images.
Since the network is trained jointly, losses are linearly combined using a factor penalizing more heavily segmentation errors.
Accuracy measured in terms of the Dice score.
Extension to multi-class problem
Extension of the segmentation subnetwork to \(K\) classes is done by by changing the number of final feature maps to match the \(K\). The segmentation loss weight factors and the classification penalization factor are modified according to empirically found factors.
Each tumor subclass has to be present at least once in each training batch.
Image-level labels are provided for each class (absent/present in the image).
Authors found better accuracies when each subclass has its dedicated entire classification branch.
Data
BraTS Challenge MRI brain datasets, doing 2D, axial slices segmentation. They assume slice-level labels for weakly-annotated images, and use 220 slices with slice-level labels and a varying number (5, 15, 30) of fully-annotated MRI slices.
T1, T2, T2+contrast and T2-FLAIR contrast images are used.
228 training images, 57 test images.
Benchmark of challenge split into three categories/tumor regions: whole tumor (all tumor subclasses), tumor core (“non-enhancing core”, “contrast-enhancing core”) and enhancing core (“contrast-enhancing core”).
Pre-processing
Skull-stripping, resampling to isotropic resolution, and registration of images to a common brain atlas, and intensity normalization.
Test setting
Three different training scenarios, with a varying number of patients with fully-annotated data (5, 15 et 30).
In each binary case, the model is trained for segmentation and classification of one tumor region.
Results
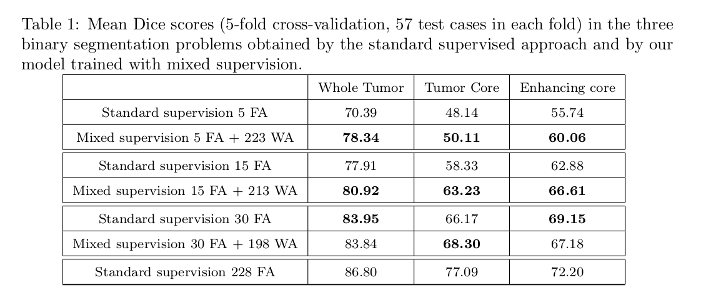
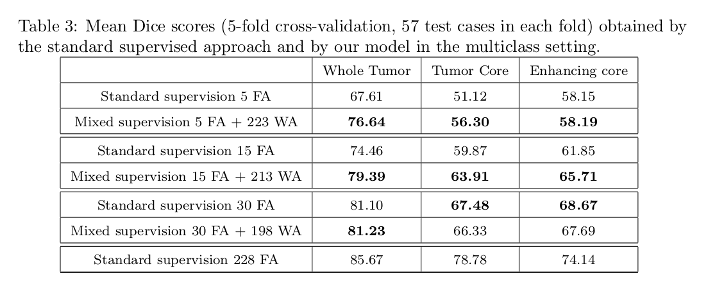
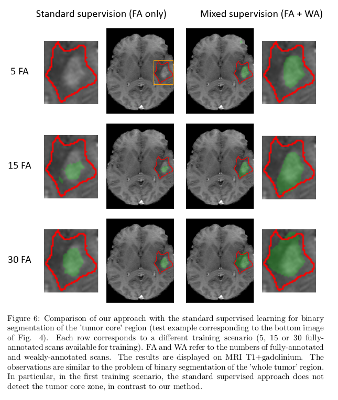
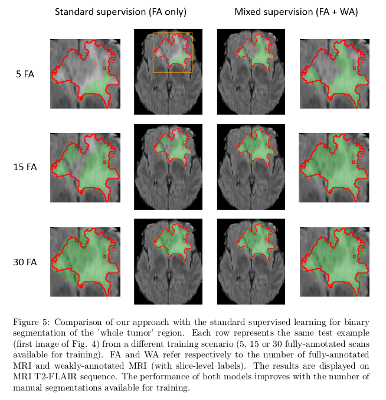
The model with mixed supervision provides a significant improvement over the standard supervised approach when the number of fully-annotated images is limited.
Segmentation performance increases quickly with the first fully-annotated cases, both for the standard supervised learning and the learning with mixed supervision.
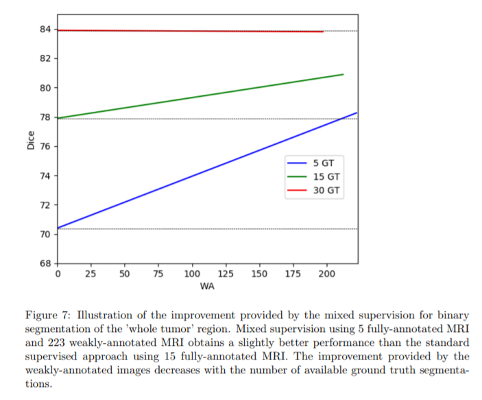
The improvement provided by the weakly-annotated images decreases with the number of available ground truth segmentations.
Dice scores tables:
Dice score improvement with increasing number of weakly annotated images used with respect to bare fully annotated image score:
Segmentation results:
Conclusions
Authors propose a neural network producing both voxelwise and image-level outputs.
Their experiments showed that the use of weakly-annotated data improves the segmentation performance significantly when the number of manually segmented images is limited.
Comments
Since the BraTS database contains only tumor images, authors needed non-tumor images to train the classification part of the model. The source of these images is not disclosed.
The trained networks variance on the sequence used for prediction is not shown.