DeepTract: A Probabilistic Deep Learning Framework for White Matter Fiber Tractography
Highlights
- Authors propose a deep-learning framework for estimation of white matter fibers orientation and streamline tractography from raw diffusion MRI data, without assuming a specific diffusion mode.
- Method to perform both deterministic and probabilistic tracking.
- Authors propose to use a discrete classification framework, rather than a regression framework to allow for probabilistic tracking.
- Improved performance scores are reported over state-of-the art classical and DL methods on the ISMRM 2015 Challenge dataset1.
Introduction
Data-driven approaches and Machine Learning (ML) techniques aim to learn the mapping between Diffusion Weighted Images (DWI) signals and fiber orientations directly from Diffusion Magnetic Resonance Imaging (dMRI) and tractography datasets, instead of using a-priori modeling.
A variety of ML and Deep Learning (DL) methods have been proposed to date. All provide a single streamline direction in each step (deterministic tractography). Also, they do not attempt to model the underlying fiber orientation distribution function (fODF), but rather directly provide a single tracking direction.
DL has also been applied to fODF reconstruction. These works, however, do not perform fiber tractography.
Authors present a DL-based tractography framework, addressing both fiber orientation estimation and streamline tracking from raw dMRI data.
Methods
Input Model
Authors train an RNN model for predicting local fiber orientations (i.e., classes) prior to tracking. Predicted local fiber orientations are assumed to be distributed according to a discrete probability distribution.
They feed their RNN with the sequence of DWI values along each streamline segment.
Fiber Orientation Estimation
Given an input sequence, the network predicts a vector of \(M + 1\) class probabilities \(P(\textbf{d})\) along each point of the input streamline (\(M\) represents the number of classes of an fODF discrete sampling over the unit sphere, to whom an “end-of-fiber” class is added).
These distributions differ from standard fODFs, since they are conditioned by the DWI “history” along a specific streamline path. They refer to them as conditional fiber orientation distribution functions (CfODFs).
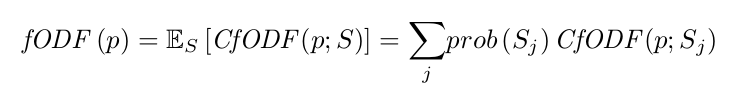
Streamline Tractography
During training, the predicted CfODFs are compared to the “true” orientation derived from input streamline itself.
To penalize larger mistakes, true ground-truth labels are convolved with a Gaussian kernel to compare the generated probability distribution.
Deterministic tracking is performed by stepping in the most likely fiber orientation, i.e. \(\hat{d}_0 = arg {max}_{d} CfODF (p_0)\). Probabilistic tracking can be performed by sampling a random direction from the CfODF.
![]()
Tracking termination
The streamline is propagated iteratively as in standard tractography algorithms according to \(\hat{p}_{k+1} = \hat{p}_{k} + \alpha \hat{d}_{k}\). The process is repeated until the “end-of-fiber” class is predicted.
To avoid accumulated errors leading to completely erroneous outputs, the tracking process of a streamline is terminated whenever the entropy of the predicted CfODF exceeds a threshold fixed according to the rule \(E_{th}(t) = a \cdot exp \left( -\frac{t}{b} \right) + c\).
Network Architecture
- Gated Recurrent Unit (GRU) RNN.
- Adam optimizer with a batch size of 40 streamlines per batch.
- Dropout was used with deletion probability of 0.3 to avoid overfitting.
- Gradient clipping to avoid exploding gradients.
- Streamline tractography on new data using a fixed step size of 0.5 (in voxels).
- Seeding: 100K randomly placed seed points (one seed per voxel at most).
- Tracking termination: high-curvature steps (larger than 60°). Entropy-based stopping criteria: \(a=3\), \(b=10\) and \(c=4.5\).
- Streamlines shorter than 20 mm or longer than 200 mm were discarded.
Data
- ISMRM 2015 Tractography Challenge phantom dataset1.
- Training on (i) ground-truth tractogram; (ii) MITK2 tractogram.
- “Ground-truth” whole brain tractography created using Q-ball reconstruction, followed by probabilistic streamline tracking using MITK diffusion tool. Resulting streamlines were randomly divided into training and validation sets. Data augmentation was performed by reversing the orientation of all streamlines in the training set.
Results
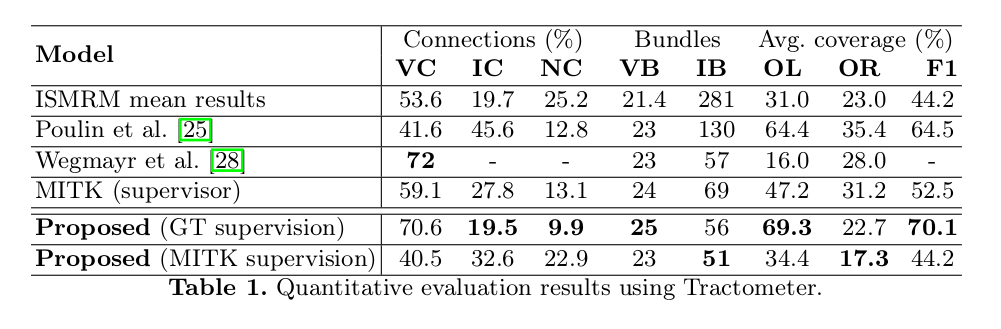
- Evaluation using the Tractometer3 score.
- Overfitting when training on ground-truth data.
- Better generalization than ISMRM 2015 Challenge contestants when training on MITK output.
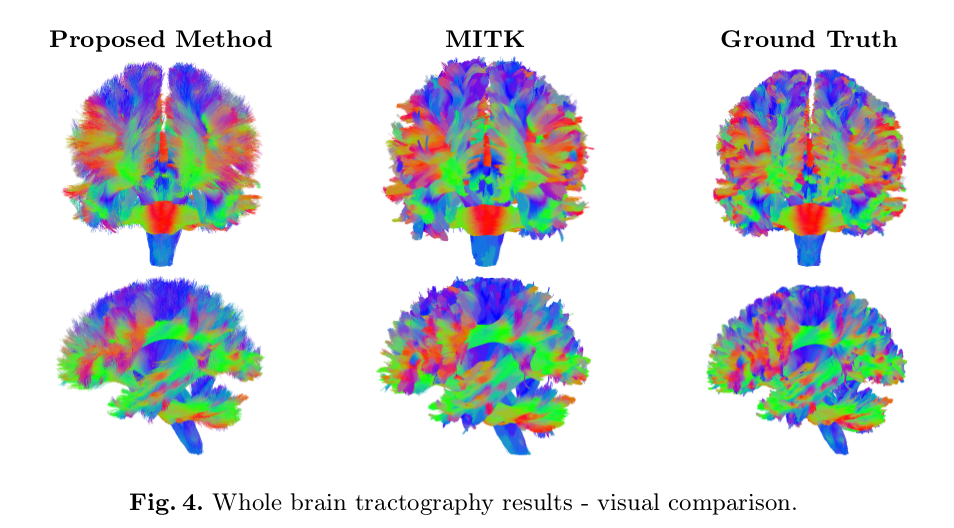
- Qualitative (visual) demonstration of bundle-specific probabilistic tractography performed shows results “are in-line with the ground truth tractograms.”
![]()
Discussion
- Deep learning framework capable of performing both deterministic and probabilistic streamline tractography directly from raw dMRI data.
- Comparable performance to state-of-the-art classical and DL-based tractography algorithms.
Comments
- Looks heavily inspired (including network architecture and tracking parameters) by Poulin et al.4.
- The way the assessment is made seems not to be a standardized way to do it, using MITK both to generate a training dataset and its results being reported in the comparison.
- Training on the ground-truth seems not a fair practice. At inference time, the model has already seen the same tracks, so it will necessarily provide good results.
- A few conceptual things to note are that:
- Their fODF, unlike the model defined in the domain, does not have any underlying model, and is composed of directions sampled from the unit sphere.
- Their probabilistic tracking is based on a sampling a random direction from the CfODF, which is different from what probabilistic tracking methods do.
References
-
Klaus Maier-Hein, Peter Neher, Jean-Christophe Houde, Marc-Alexandre Côté, Eleftherios Garyfallidis, Jidan Zhong, Maxime Chamberland, Fang-Cheng Yeh, Ying Chia Lin, Qing Ji, et al. Tractography-based connectomes are dominated by false-positive connections. biorxiv, 084137, 2016. ↩ ↩2
-
Marc-Alexandre Côté, Gabriel Girard, Arnaud Boré, Eleftherios Garyfallidis, Jean-Christophe Houde, and Maxime Descoteaux. Tractometer: towards validation of tractography pipelines. Medical image analysis, 17(7):844-857, 2013. ↩
-
Philippe Poulin, Marc-Alexandre Côté, Jean-Christophe Houde, Laurent Petit, Peter F Neher, Klaus H Maier-Hein, Hugo Larochelle, and Maxime Descoteaux. Learn to track: Deep learning for tractography. In International Conference on Medical Image Computing and Computer-Assisted Intervention, 540–547. Springer, 2017. ↩