NeuroNet: Fast and Robust Reproduction of Multiple Brain Image Segmentation Pipelines
Highlights
- NeuroNet is a deep convolutional neural network mimicking multiple popular and state-of-the-art brain segmentation tools including FSL1, SPM2, and MALPEM3.
- Produces outputs from raw images, avoiding pre-processing steps (e.g. bias correction or skull-stripping).
- Reduced processing time by an order of magnitude compared to running each individual software package.
- Good reproducibility of the original outputs.
Introduction
The disparity of results in brain tissue segmentation in neuroimaging tools such as FSL1, SPM2, and MALPEM3 puts neuroscientists before the dilemma of which tool to choose.
Authors hypothesize that learning a CNN model from multiple gold-standard reference segmentations would allow to wash out biases and errors.
Also, learning jointly from hierarchical sets of class labels should have the potential to increase the overall accuracy based on theory derived from multi-task learning.
Methods
- An autoencoder is used based on a ResNet encoder and multiple FCN decoders, as many as the number \(k\) of output segmentations coming from different tools.
- Multitask loss: average categorical cross-entropy.
- Each data is automatically segmented using three different state-of-the-art tools (FSL, SPM and MALP-EM) generating five outputs in total. The automatically generated label maps from the three tools serve as training data for NeuroNet.
- For structures that are present in the output of all three tools such as white matter (WM), gray matter (GM) or cerebrospinal fluid (CSF), three different estimates are obtained (one from each tool) which are all used as gold-standard reference.
- Compared NeuroNet variants include training
- on single targets
- on all targets
- on tissue segmentations only
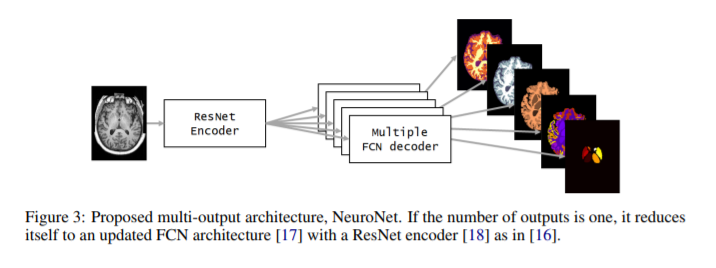
Data
Trained on \(5000\) T1-weighted brain MRI scans from the UK Biobank Imaging Study4 with data augmentation; \(10\) validation volumes; and \(713\) test datasets.
Pre-processing
- Input volume size homogenized to \(128 \times 128 \times 128\) voxels.
- Input volume images normalized to zero mean and unit standard deviation using volume statistics.
Results
- Comparison metric: Dice Similarity Coefficient (DSC).
- Multi-output architectures yielded slightly reduced mean DSC over training several single networks. May be due to having fixed hyperparameters across experiments.
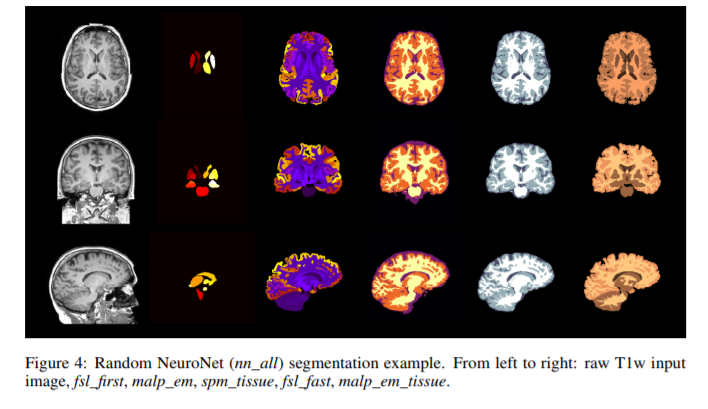
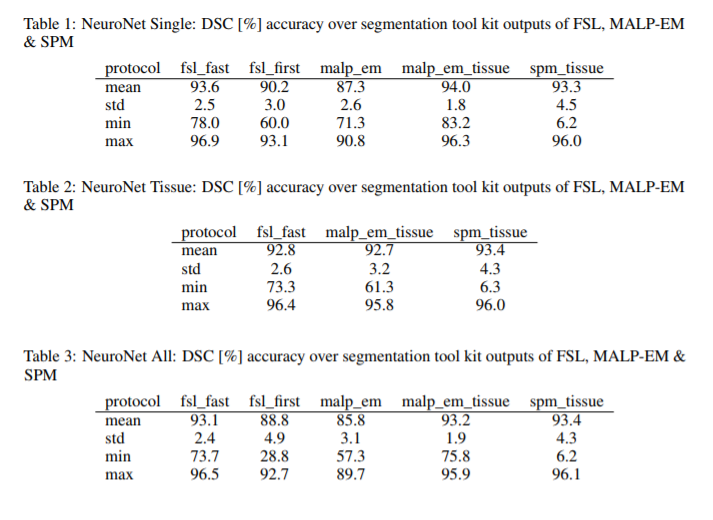
Conclusions
- Trained a single model from these complementary and partially overlapping label maps.
- Powerful “all-in-one”, multi-output segmentation tool.
- The method reduces inference time with respect to the state-of-the-art tools.
Comments
- Batch size of 1 seems not to be a batch.
- Data augmentation procedure (“random crops of size \(128^3\)”) seems unclear.
- Finally, the proposal of using the whole collection of data for training the model does not compare well, so it deserves a more fair comparison.