Unsupervised brain lesion segmentation from MRI using a convolutional autoencoder
Summary
Problem: segment brain lesions with an unsupervised deep neural net. The main difficulty is that lesions can be anywhere, have any shape and any size.
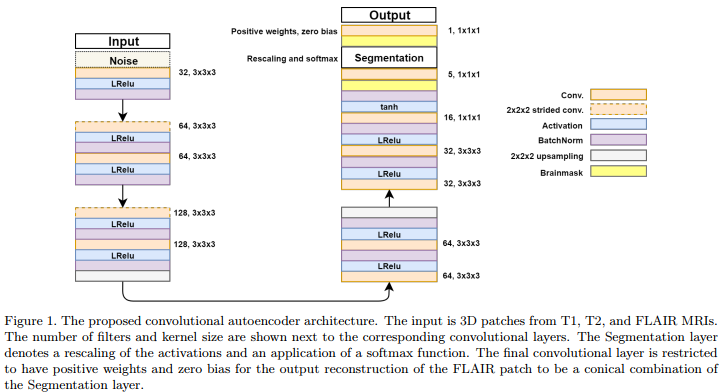
Solution: use a convolutional autoencoder which incorporate a “segmentation layer” before the output. (c.f.Fig.1)
Specific details
Due to space limitation, the authors process a brain patch-wise, each patch being a 80x80x80 volume. For training, patches have a stride of 40 while at test time they use a stride of 20 (with an average of overlapping voxels). The use the following reconstruction loss :
\[L = (Y^p - \hat{Y}^p)^2\]with p=3. A brainmask is applied before the loss so the background is not considered. Furthermore, Gaussian noise with zero mean and a standard deviation of 0.05 was added to the input during training for noise robustness.
And last, the reason for having a “segmentation” layer before the output is explained as follows :
The final convolutional layer is restricted to have positive weights and zero bias for the output reconstruction of the FLAIR patch to be a conical combination of the Segmentation layer.
Experiments
Dataset:
- 4811 brain MRI, all with T1, T2 and FLAIR.