Learning Implicit Brain MRI Manifolds with Deep Learning
Highlights
Implicit manifold learning of brain MRI through two common image processing tasks:
- Unsupervised synthesis of T1-weighted brain MRI using a Generative Adversarial Network (GAN) by learning from 528 examples of 2D axial slices of brain MRI.
- U-net autoencoder with layer sums for image denoising showing higher PSNR than FSL SUSAN after denoising.
Introduction
The work assumes that a low-dimensional, non-linear manifold can effectively represent the variability found in brain anatomy.
The mapping of a brain image into a manifold and vice versa can help construct priors that span the entire variability spectrum of a normal human brain.
Authors hypothesize that the ideal manifold mapping would learn a naive representation using the entire image and from this manifold generate images that are of comparable quality as the learning set.
Previous image synthesis works have produced “near-realistic” images, but “have not been validated for image quality by imaging experts” according to the authors of this paper.
Methods
## Image synthesis
- Generator and discriminator inspired from the Deep Convolutional Generative Adversarial Network (DCGAN) architecture.
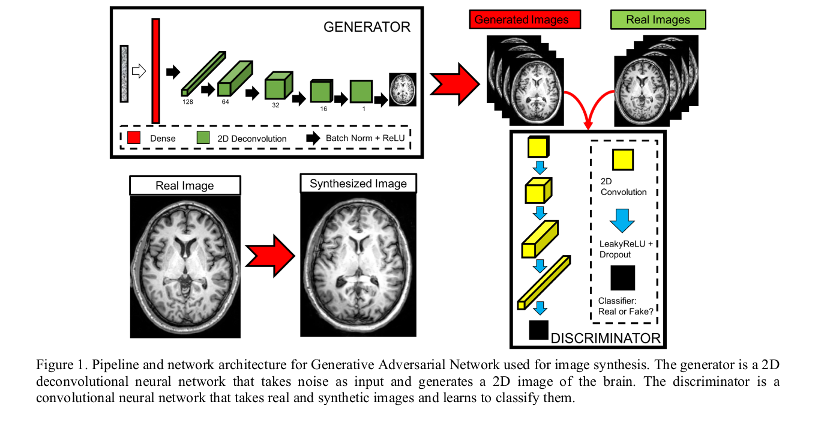
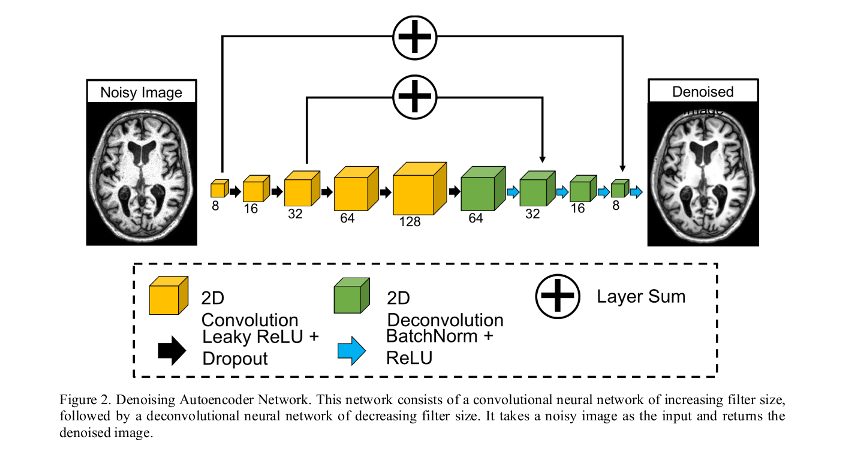
## Image denoising
- Features “symmetric skip connections”: adds the feature output from 2 conv layers to the corresponding deconv layer.
- Training set: 528 noise vectors generated by adding Gaussian noise within a mask of the brain and skull to the 2D slices of the dataset.
- Testing set: 100 new noise vectors.
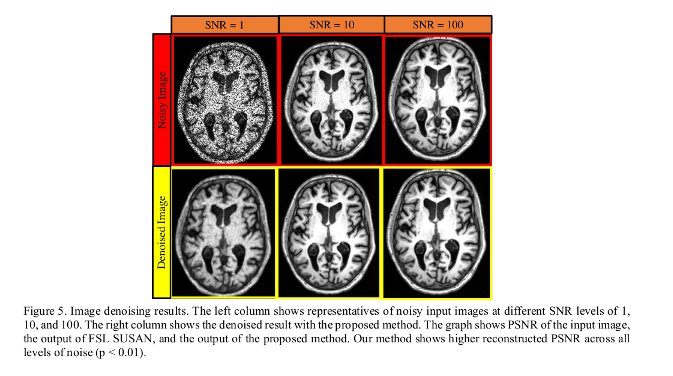
- Three different noisy images were generated for each subject with SNR of 1, 10, and 100. Three separate instances of the proposed autoencoder network were trained for each level of noise.
- Minimize the MSE.
Data
- 528 T1-weighted brain MPRAGE MRI images (single mid-line axial slice) of healthy subjects from the Baltimore Longitudinal Study of Aging (BLSA).
- \(220 \times 172\) size images with \(1\) mm isotropic pixel resolution.
- Affine-registered to MNIs-space and intensity-normalized.
Results
Image synthesis
- Synthesized images were shown to be unique by performing a cross-correlation with the training set.
- Real images with high correlation show several structural differences from the synthesized images, such as ventricle size or gyral patterns, suggesting that this network is not simply reconstructing examples from the training set.
- “Average quality” score by two separate raters: intra-rater overlap in the distribution score histogram.
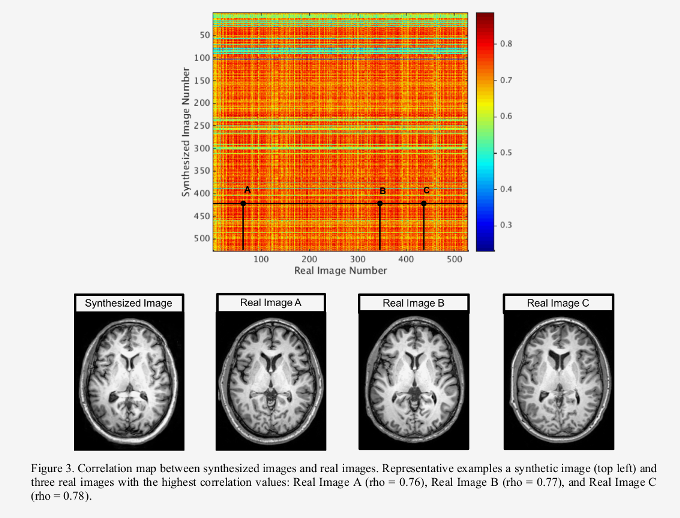
Image Denoising
Discussion
. Generated images are rated with comparable quality to the real images.
- Used only 528 images.
Conclusions
- Anatomic accuracy could be attained by imposing structural restrictions, such as the inter-layer connections used in the autoencoder or symmetry connections within layers.
- Denoising method is superior in terms of PSNR to the SUSAN method.
- Results may be improved by using whole 3D volume datasets and a wider population sample (e.g. including disease subjects, different acquisition equipment, sequences and parameters, etc.).
- Using image priors could allow for style transfer-like inter-modality image synthesis, such as CT dataset generation from MRI datasets.
- No image analysis experiment is performed to demonstrate that the proposed methods provide “quantitatively similar” results.