Mapping connectomes with diffusion MRI: deterministic or probabilistic tractography?
Summary
To study brain connectivity, the preferred approach is to do whole-brain tractography, then to cluster the generated streamlines, or “connections”, into meaningful groups. This is usually done using a pre-computed brain parcellation (a partition of the cortex surface).
However, there are some problems when doing whole-brain tractography, namely:
- False positive connections (implausible connections, due to e.g. bottlenecks)
- Gyral biases (streamlines mostly connect at the bottom of the gyrus instead of uniformly)
The goal of the paper is to study which family of tractography algorithms is best suited for connectome analysis: deterministic or probabilistic?
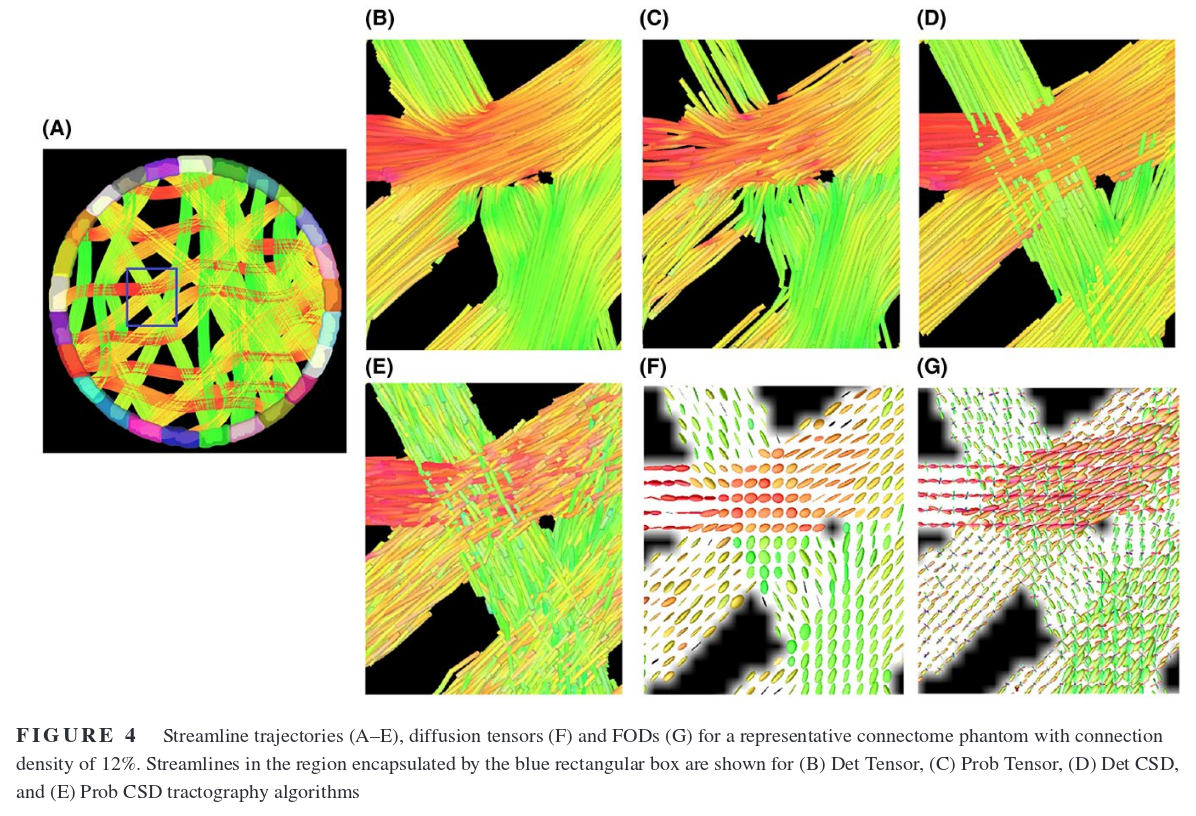
Reminder: Deterministic vs probabilistic tractography
Deterministic tractography chooses the fODF peak nearest to the previous direction
Probabilistic tractography samples from the fODF restrained to an aperture cone aligned to the previous direction
Experiments
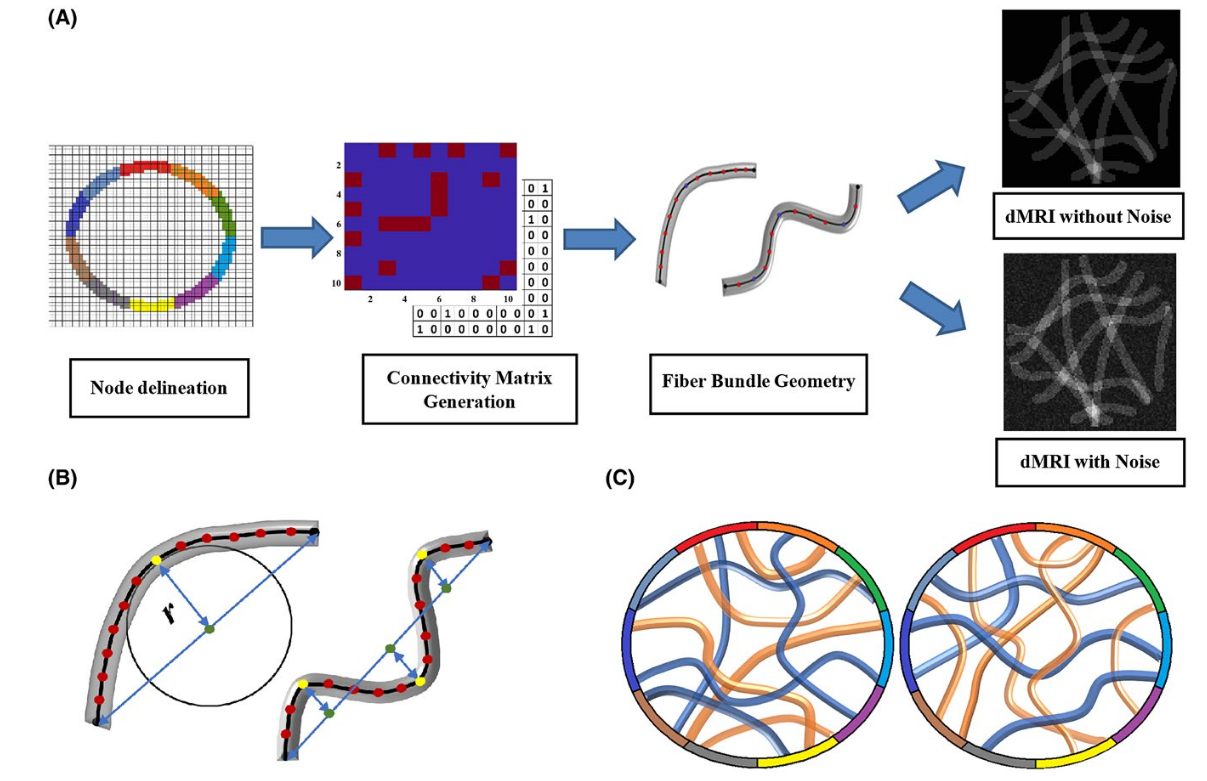
The authors used simulated phantoms (artificial “white matter” configurations) to compare deterministic and probabilistic tractography.
Results
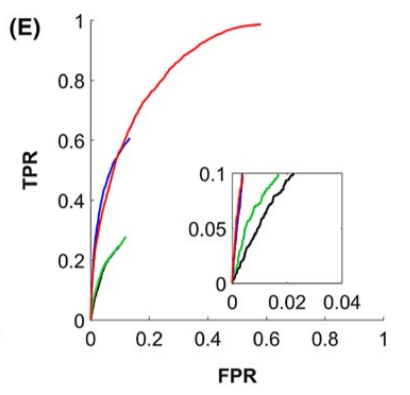
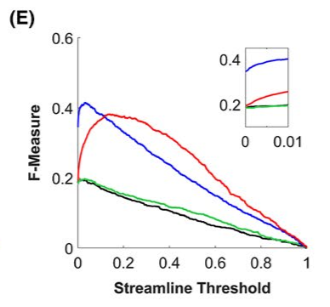
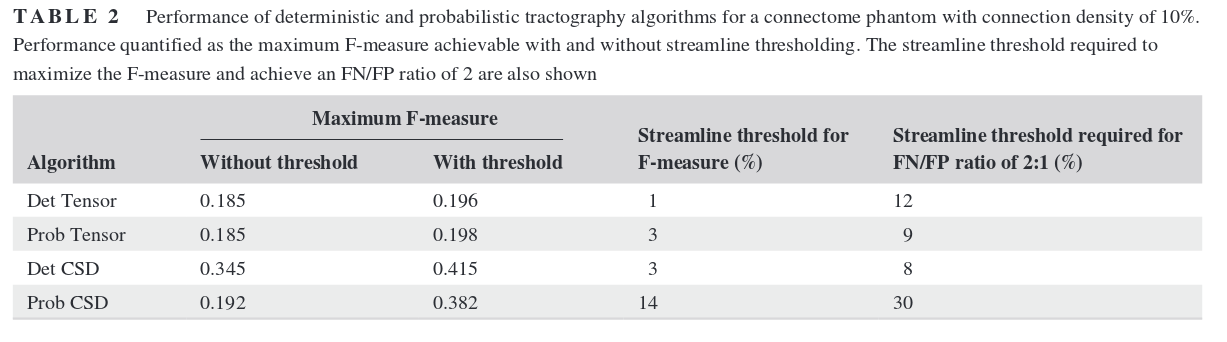
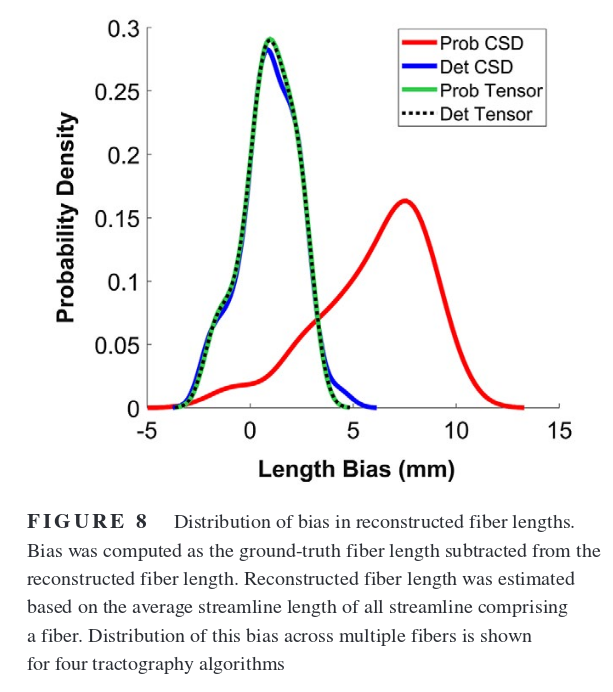
Deterministic is better because it produces fewer false positives. Probabilistic algorithms would be better suited for tractometry or other single-bundle analysis. Thresholding streamlines to remove the “bundles” with the lowest number of connections helps, but the threshold needs to be tuned, and the ideal value varies a lot depending on the algorithm used or the connectivity of the phantom…