Exclusive Independent Probability Estimation using Deep 3D Fully Convolutional DenseNets for IsoIntense Infant Brain MRI Segmentation
Highlights
- 3D FC-DenseNet architecture, a multi-label patch-wise training technique with balanced similarity loss functions and a patch prediction fusion strategy for segmentation.
- Focus on training part.
Summary
The goal is to extract the white matter (WM), gray matter (GM) and cerebrospinal fluid (CSF) in infant brain MRIs.
The proposed method segments the mutually exclusive brain tissues with similarity loss function parameters that are balanced based on class prevalence.
Introduction
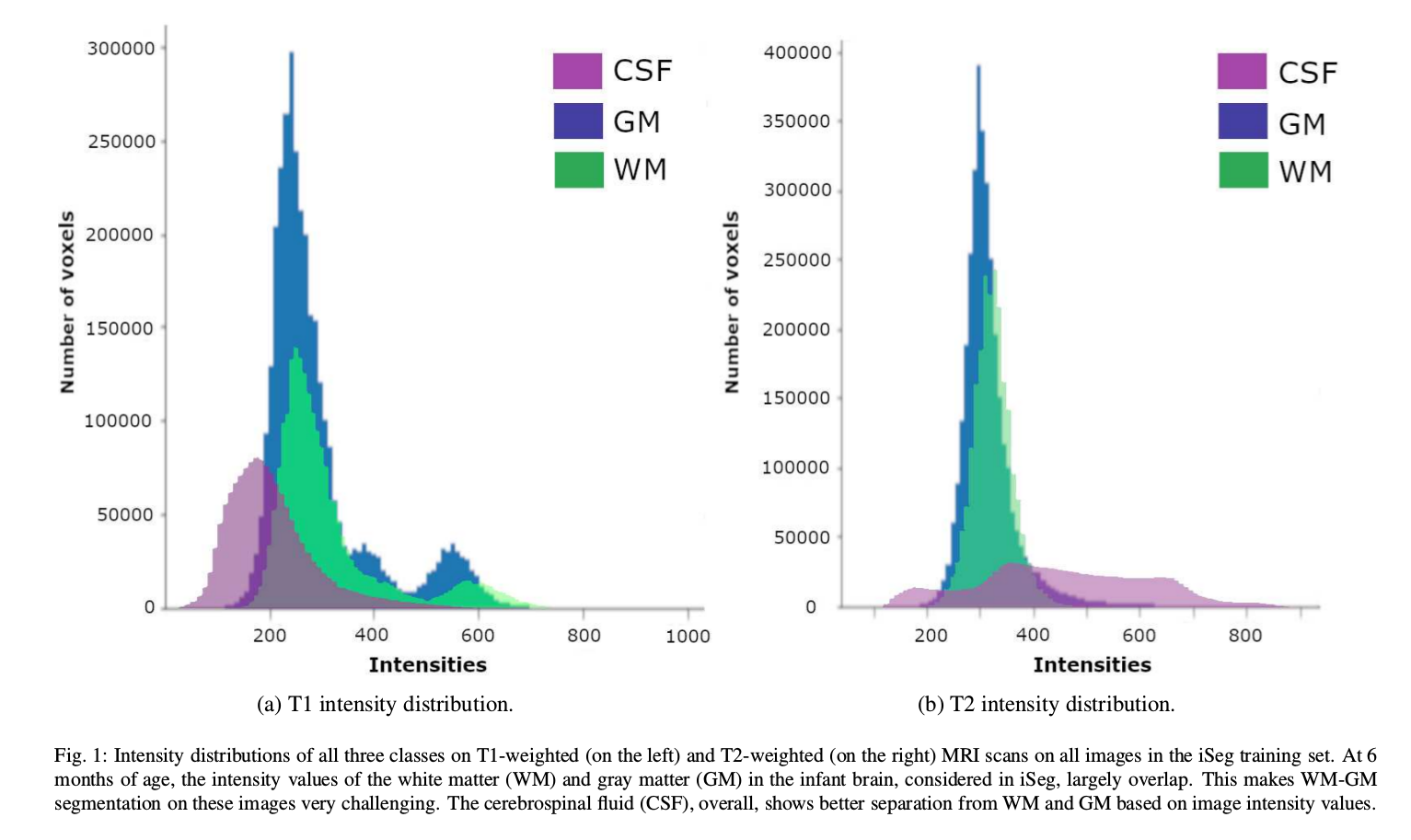
- White matter (WM) and gray matter (GM) of the developing brain at about 6 months of age show similar T1 and T2 relaxation times: have similar intensity values on both T1- and T2-weighted MRI scans (iso-intensity).
- Most widely adopted architecture to date: DenseNets using conventional training strategies based on cross-entropy loss function.
Methods
- Architecture
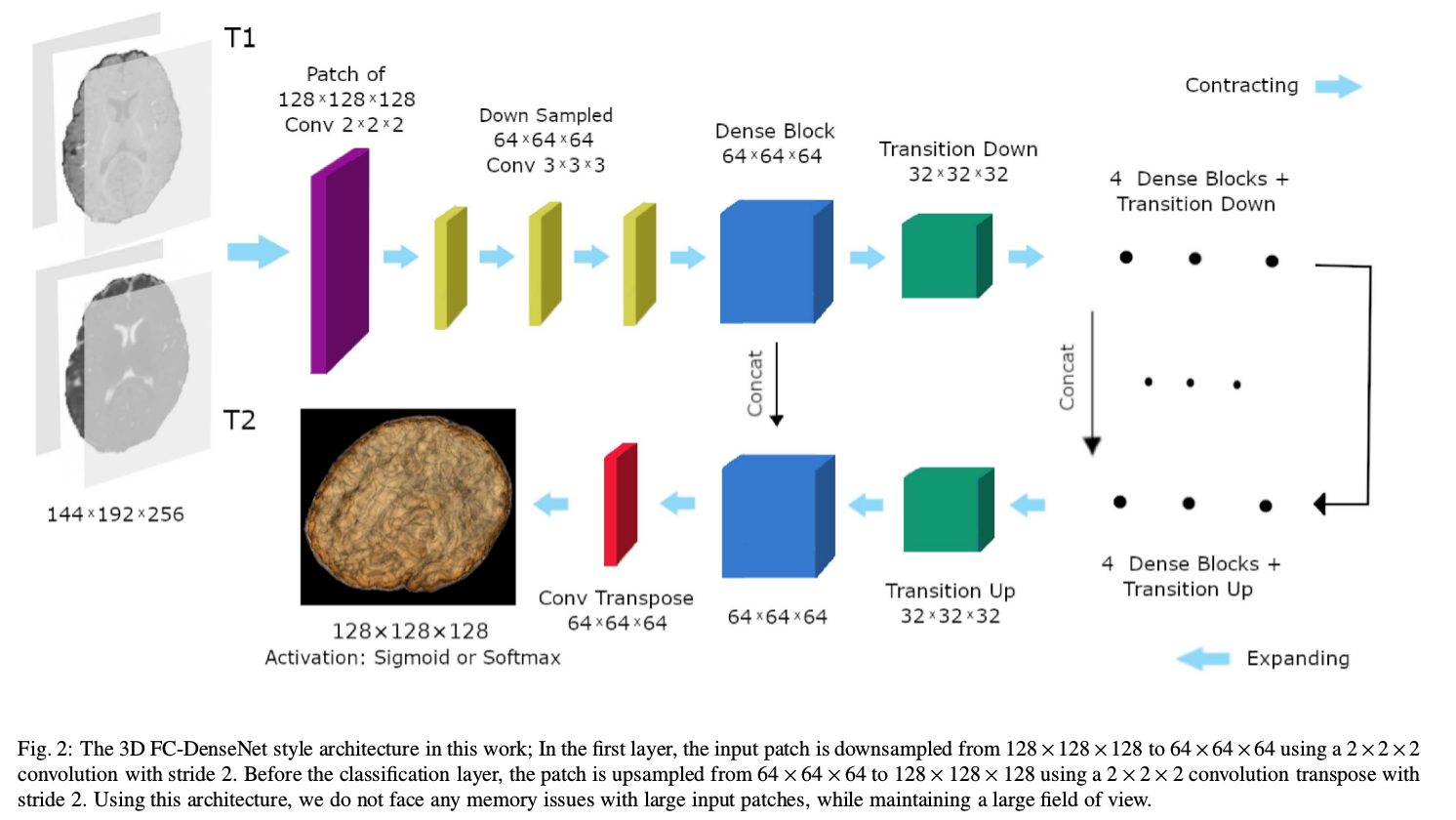
Network
- Two-channel (T1 and T2) fully convolutional (FC) DenseNet.
- Dense blocks: bottlenecks + conv layers; skip connections between layers inside dense blocks; max pooling (in transition down blocks). All conv layers followed by batch normalization and ReLU non-linear layers. Dropout used after $3 × 3 × 3$ conv layers of each dense block.
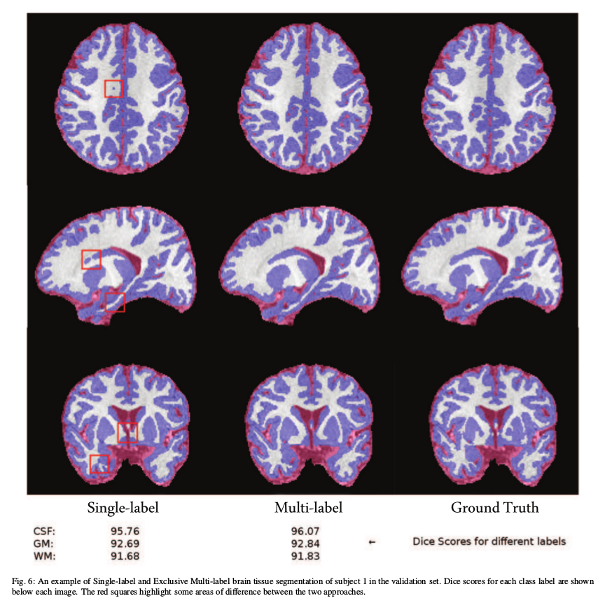
- Similarity issues overcome by training on one of the isointense class labels
(WM) instead of both (exclusive multi-label multi-class).
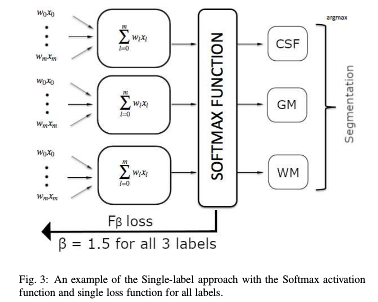
- Single-label multi-class training (each voxel can only have one label). Softmax activation.
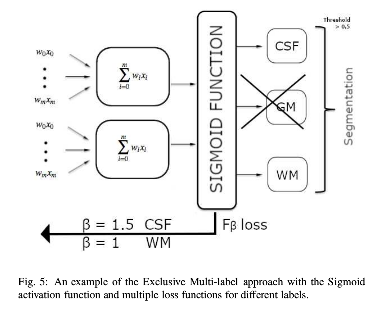
- Exclusive multi-label multi-class training (each voxel has the potential to have multiple labels). Remove GM label from training. Conclude it from the compliment of the CSF and WM labels. Sigmoid activation.
- Precision (\(TP/(TP+FP)\)) and recall (sensitivity: \(TP/(TP+FN)\)) separately balanced for each class using \(F_{\beta}\) loss functions with$ \(\beta\) values adjusted based on class prevalence in the training set.
 |
 |
- Similarity loss function:
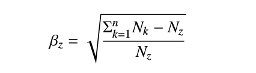
- Score:
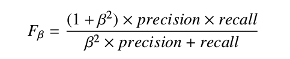
Settings
- No pre-processing (already skull-stripped and registered)
- Normalization to unit mean
- 60 layers
- Adam optimizer
- Learning rate with decay steps
- 5-fold cross-validation in training
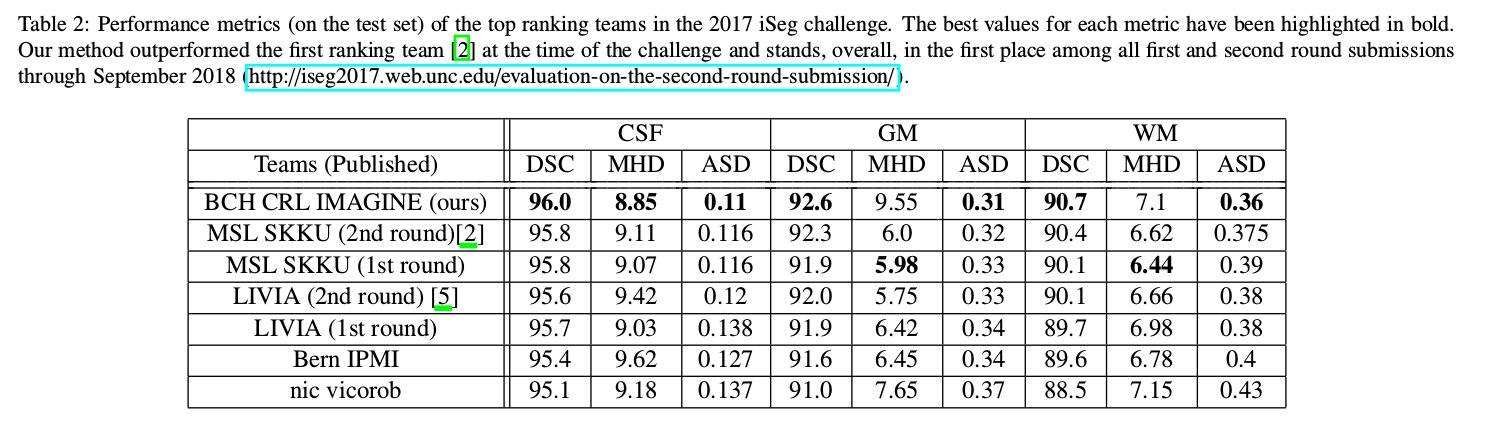
Metrics
- Dice Similarity Coefficient (DSC)
- Modified Hausdorff Distance (MHD)
- Average Surface Distance (ASD)
Data and Benchmarking
- 2017 MICCAI iSeg challenge dataset
- T1- and T2-weighed MRIs of 10 subjects
- Data augmentation: use patches on input images; rotated patches
- Prediction fusion: soft voting approach
Results
- Best results with exclusive multi-label multi-class training
- Less parameters than other contestants
- Training: 14 hours
- Testing: 90 seconds