Shallow vs deep learning architectures for white matter lesion segmentation in the early stages of multiple sclerosis
Highlights
- Compare a kNN and a CNN for MS lesion segmentation.
- CNN presents lower lesion-wise FPR, but kNN gives better Dice coefficient and volume difference.
- The combination of the two improves the results.
- The quality of the automatic segmentation is not comparable to a manual segmentation, though.
Summary
The goal is to compare a shallow and a deep learning architectures for the automated segmentation of white matter lesions in MR images of early stage multiple sclerosis patients. A combination of the two methods is finally evaluated.
Introduction
Multiple Sclerosis (MS) is a demyelinating disease that affects the central nervous system (CNS). Demyelination results in focal lesions that appear with higher frequency in the white matter (WM). ! Magnetic Resonance Imaging (MRI) is a fundamental tool for MS diagnosis and monitoring of disease evolution as well as response to therapy. Expert’s manual annotations are considered the clinical gold standard for MS lesion identification.
Methods
- Method 1: a supervised k-NN method combined with partial volume (PV) modeling. LeMan-PV is a Bayesian PV estimation (PVE) algorithm, where spatial constraints for GM and lesions are included to drive the segmentation. The spatial constraint for GM is an atlas-based probability map, and spatial constraints for lesions are derived from a kNN-supervised-based approach.
- Method 2: two 3D patch-wise ConvNet. The two networks have the same architecture and number of parameters, but don’t share the same weights. Each convolutional layer is followed by a ReLU activation function and a batch normalization regularization. Dropout (\(p=0.5\)) is applied before the first fully-connected layer. Adadelta optimizer used during training.
- Method 3: combination of the two.
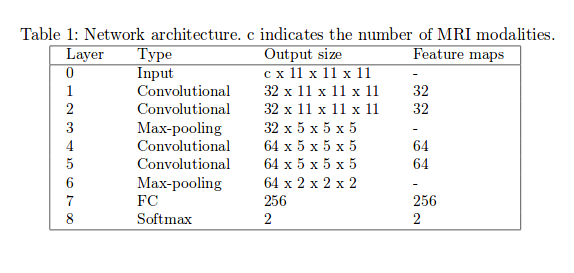
Data
All methods were trained on 32 patients (25 training set; 7 validation set), and the evaluation was performed on a test set of 73 cases.
Labels: manual segmentation by experts for the training sets; semi-automated method with manual refinement for the test cases.
Pre-processing
Metrics
- Overlap Dice coefficient (Dice)
- Lesion-wise false positive rate (LFPR)
- Lesion-wise true positive rate (LTPR)
- Voxel-wise true positives (TP)
- Volume difference (VD)
Results
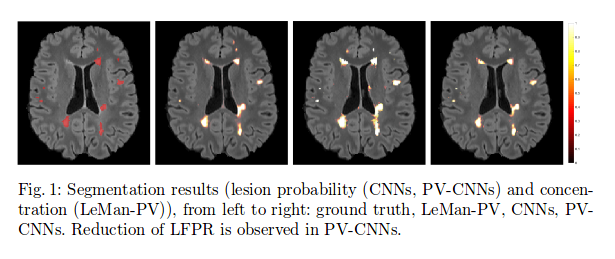
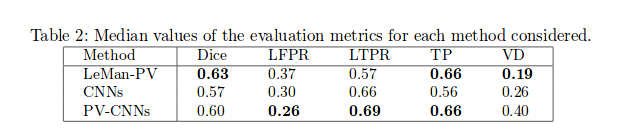
Low lesion-wise false positives (30%) for the deep learning architecture, whereas the shallow architecture yields the best Dice coefficient (63%) and volume difference (19%).
Dependence of LeMan-PV performance on the minimum lesion size considered, whereas the CNNs did not show this behavior.
Providing the CNNs with the probability maps of the LeMan-PV improved the LFPR 26%) and LTPR (69%) but did not perform well in terms of VD.
The hybrid of the two methods is also effective for WM lesion segmentation of early stages disease cases. However, further improvements are needed to increase the segmentation accuracy of low lesion burden cases, in which these automated methods achieved the worst performance (median Dice around 0.5).
Final notes
Code was largely inspired by the network architecture published here and available here.