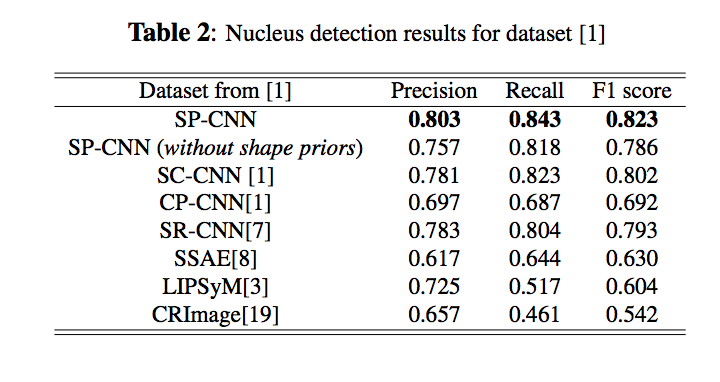
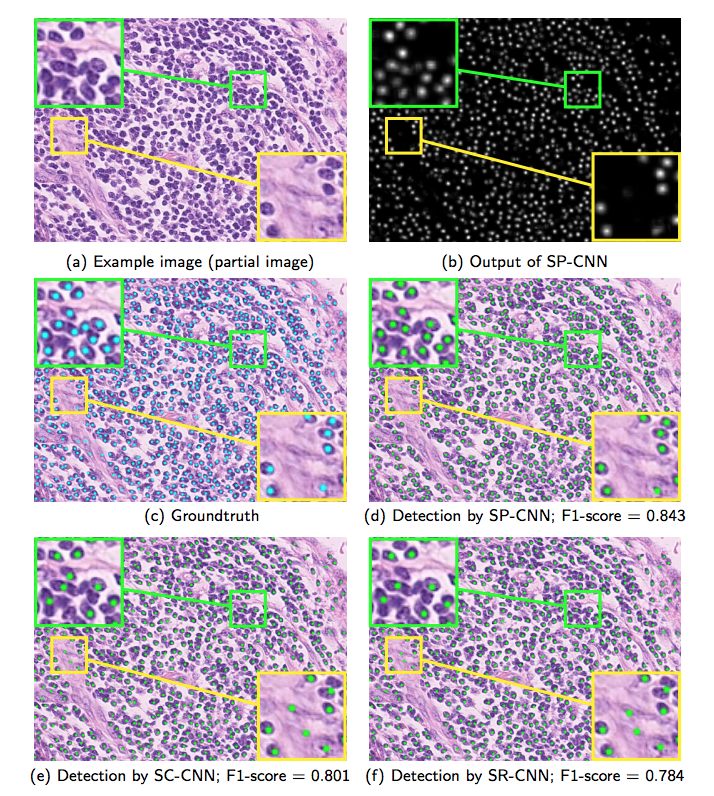
Deep Networks With Shape Priors for Nucleus Detection
Description
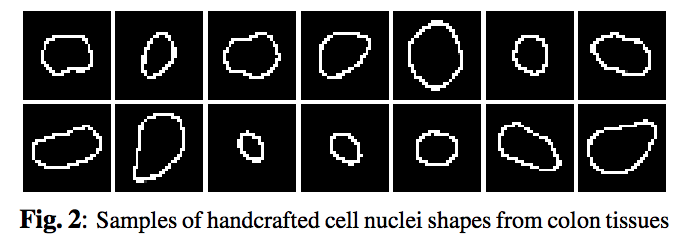
This paper proposes a network structure that can incorporate ‘expected behavior’ of nucleus shapes via two components: learnable layers that perform the nucleus detection and a fixed processing part that guides the learning with prior information which can be obtained in consultation with a medical domain expert. A set of shape priors developed with the help of domain expert to hand label the nuclei boundaries are shown in fig2. Ideally, the network’s segmentation prediction should lie inside of these nuclei boundaries (prior shapes).
Three sources contribute to generating the prior: network output, raw edge map from the input image $hat{x}$ which is obtained by Canny edge detection filter, and a set of predefined shapes $S$ prepared by the medical expert.
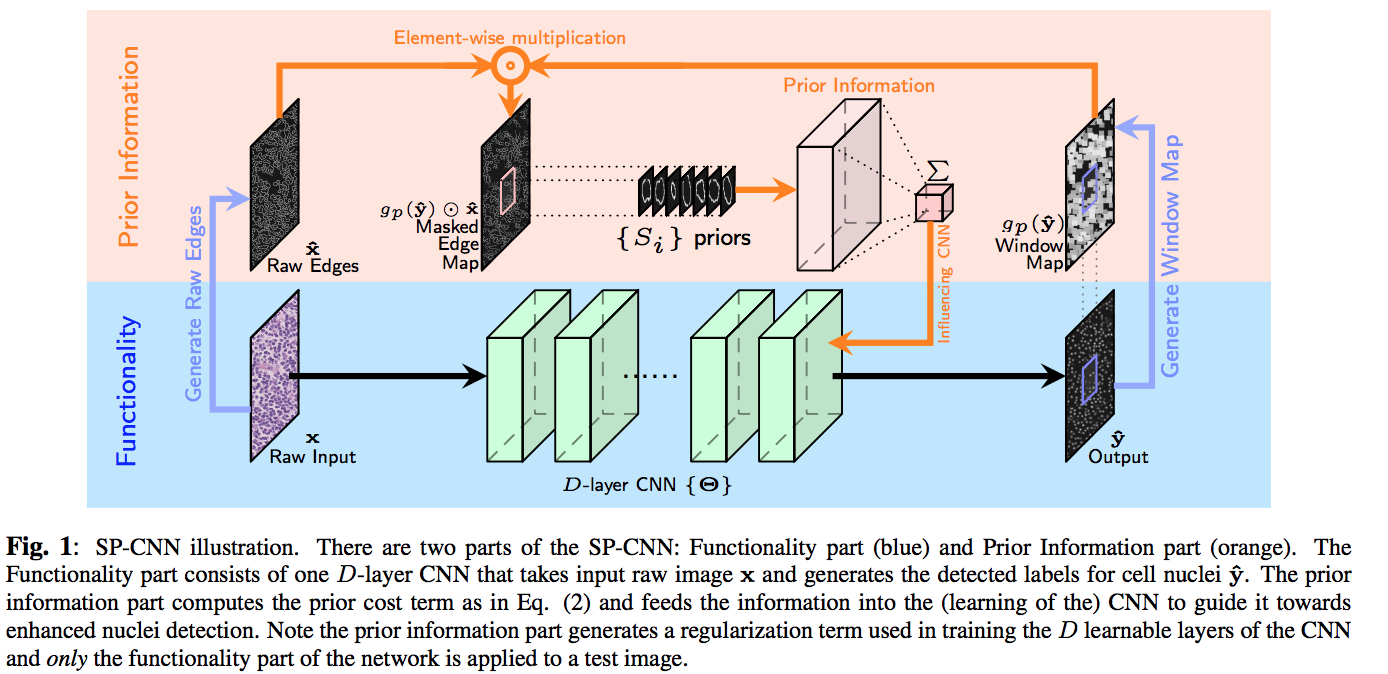
The contribution of shape prior term contains three-step (the orange part in fig1). First, the output of network max pooled with the stride of 1 and the ‘same’ padding, then it is multiplied with the raw edge image element-wise. Finally, $(gp(\hat{y}\odot \hat{x})$ The masked edge image is convolved with the shape priors in set S to generate a measurement of how well does the detection fit inside the nucleus shape.
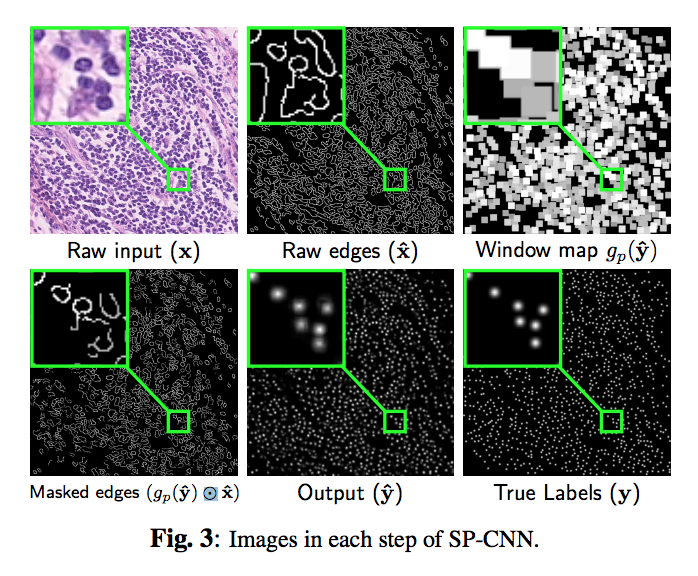
Note that the effect of the shape prior is captured by a negative regularization term since the goal is to maximize (and not minimize) correlation with ‘expected shapes’.
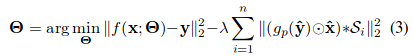
Results