Anatomical Priors for Unsupervised Biomedical Segmentation
The authors propose to use autoencoders to capture shape priors of the brain on a given dataset in order to segment another unlabeled set : “The first zero-shot neural network based segmentation of the brain”.
Segmentation pipeline
-
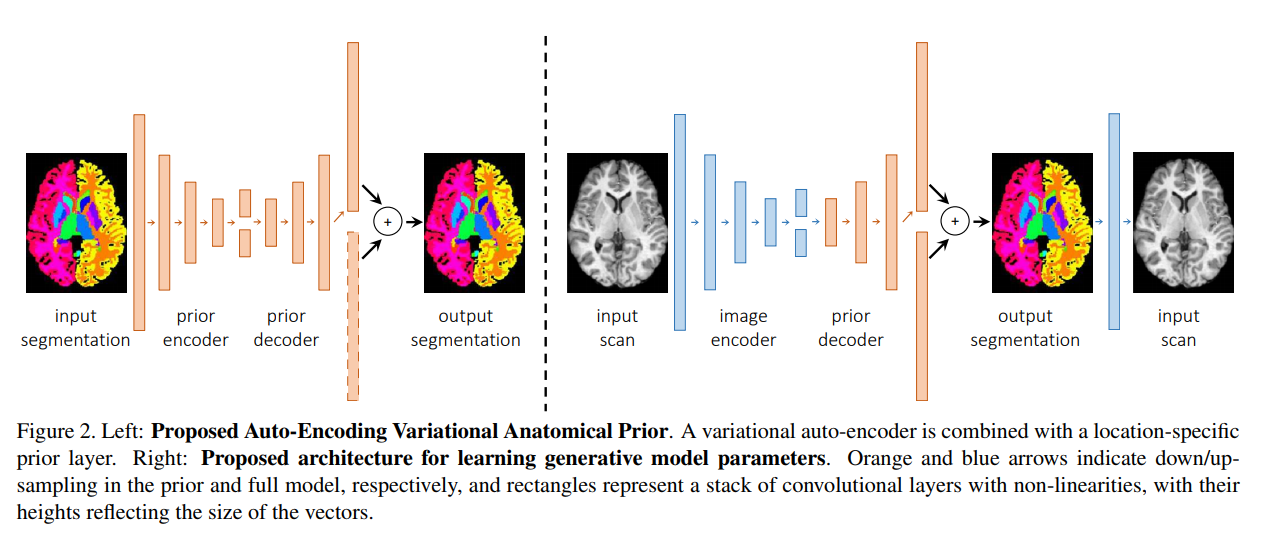
They train an autoencoder to encode shape priors from a labeled dataset as latent representations z. A location probabilistic prior is combined at the end of the decoder to denoise the reconstruction.
-
They freeze the decoding part as the decoder of a second autoencoder that takes as input images of another modality for which there is no groundtruth. They train the encoding part to optimize the reconstruction of the scan from the segmentation obtained by the decoder.
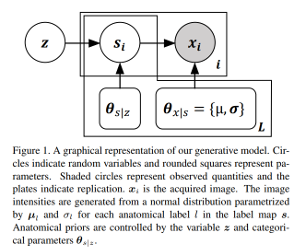
Shapes are represented as possible deformations of an atlas and their encoders learn to predict both mean latent representations (highest probability) and the variance (uncertainty). The groundtruth distributions characteristics were obtained with a generator that makes the approximation that shapes follow a gaussian distribution to make it tractable:
Experiments
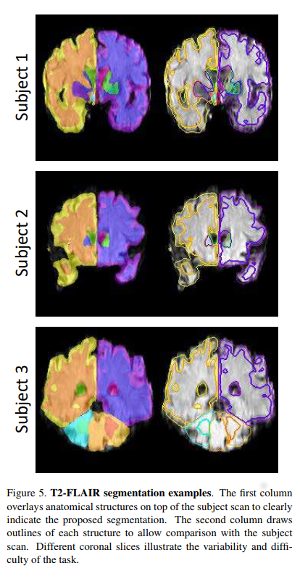
They apply this model on : Labeled dataset : 14,000 T1-weighted brain MRI scans from eight publicly available datasets. Unlabeled dataset : 3800 T2-FLAIR scans
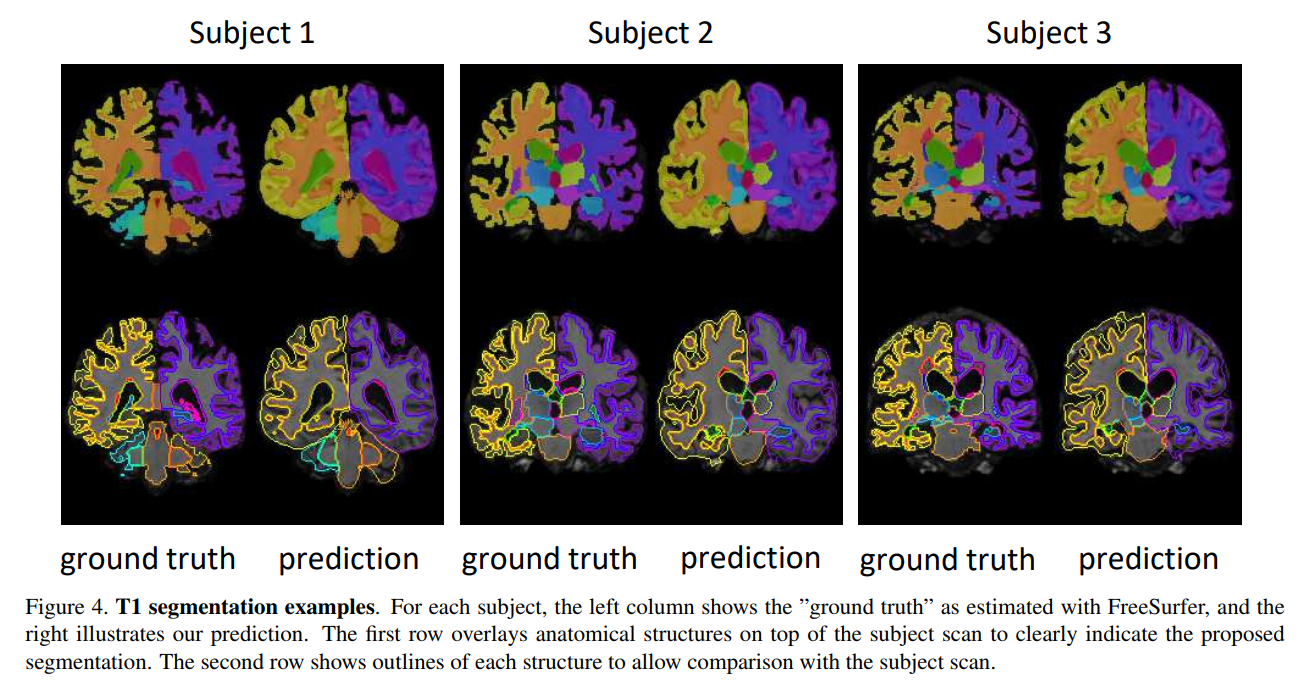
Though the Dice metric can be used for the supervised segmentation, only visual assessment is proposed for the unsupervised segmentation.