An Unsupervised Learning Model for Deformable Medical Image Registration
The goal is to train a CNN for deformable, pairwise 3D medical image registration.
Highlights
- Deformation function is learned over a dataset of images, instead of doing it for each image pairs (like ANTs).
- Atlas-based registration
- Loss function is a cross-correlation, easy to optimize on GPU
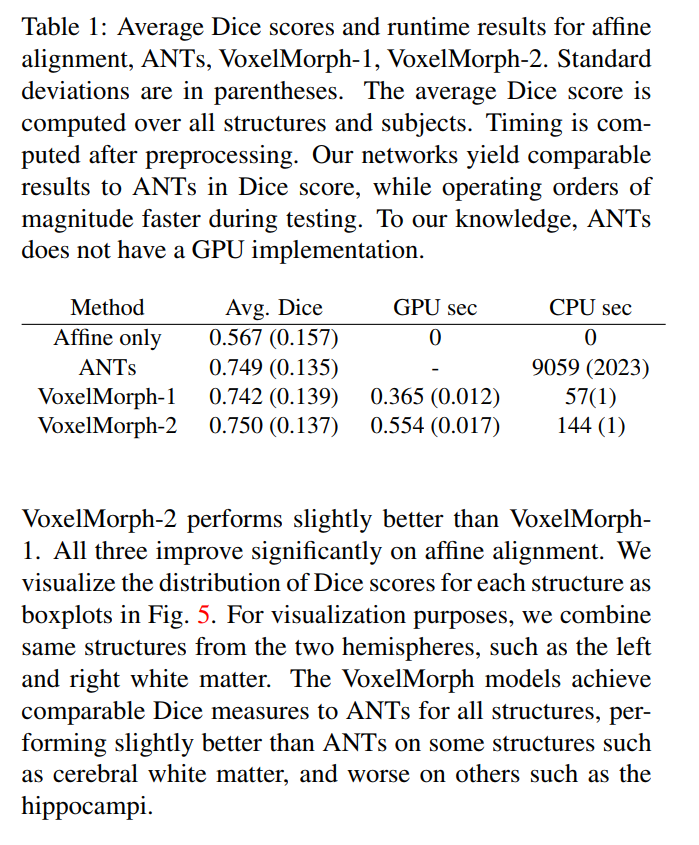
- Dice scores comparable to ANTs, but runs 130 times faster
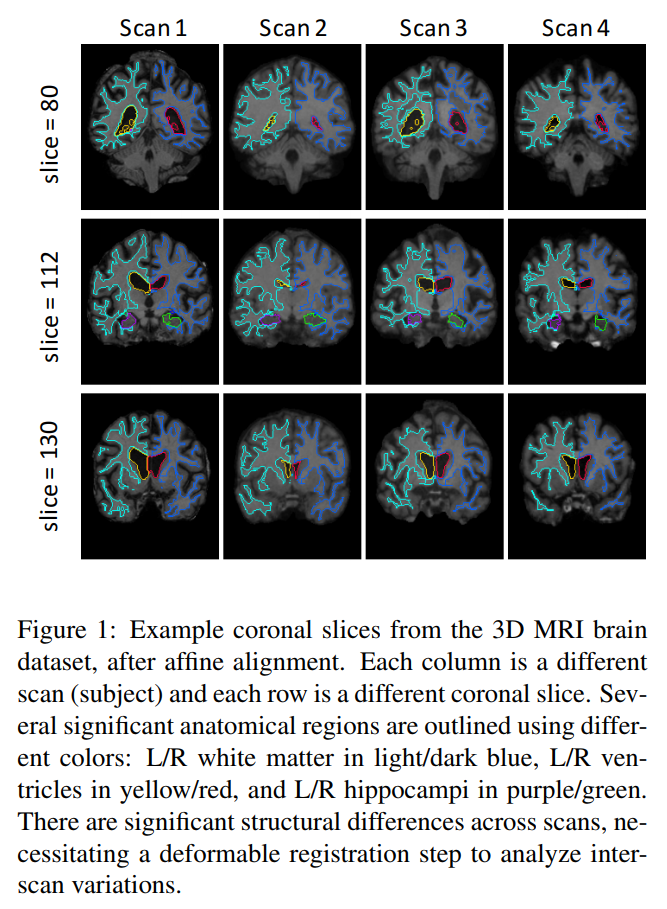
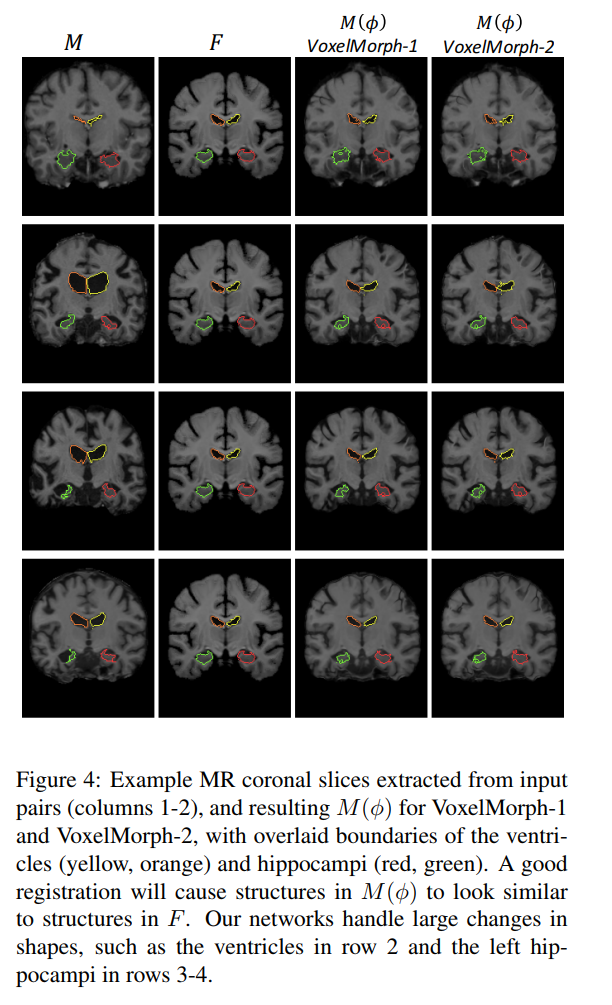
Typical images with ground truth segmentation:
Summary
-
Input pair of images are already affinely aligned (only source of misalignment is nonlinear).
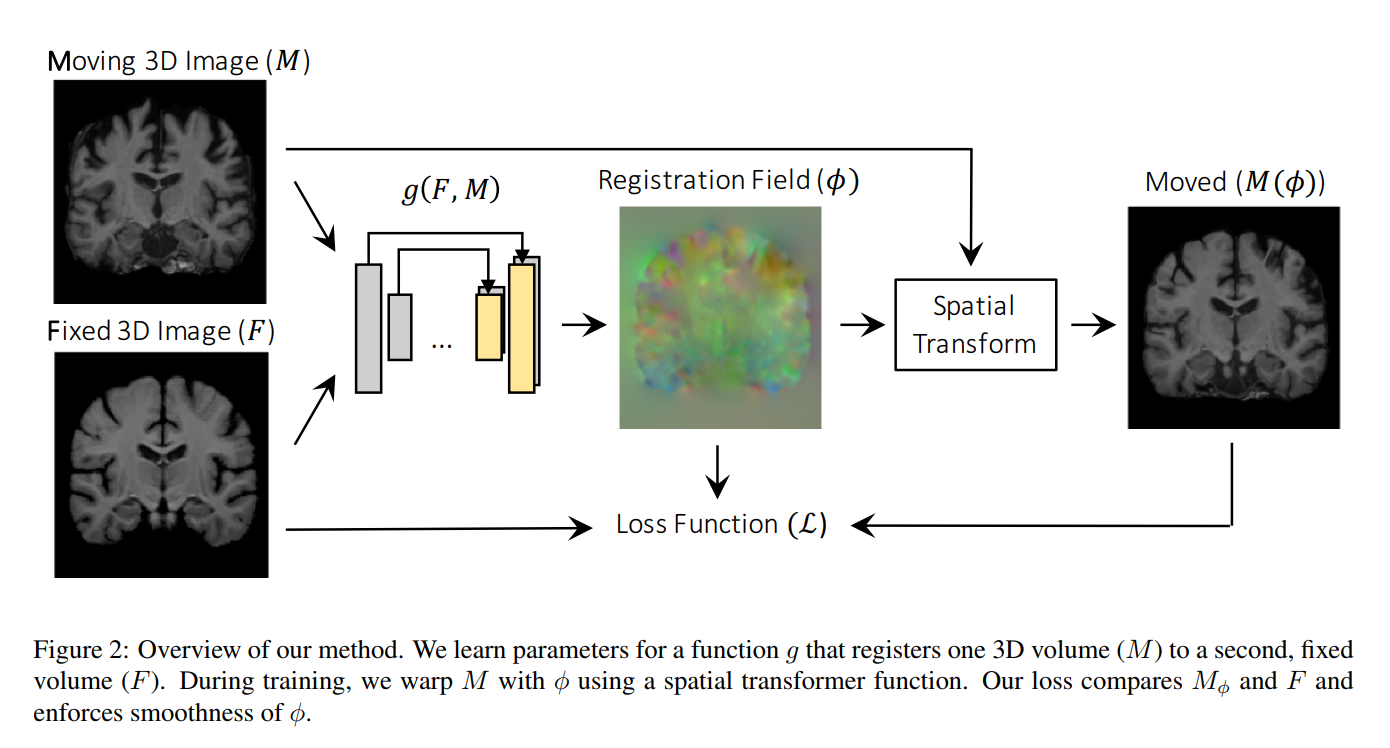
- U-Net model
- Input is 2 volumes concatenated as a 2-channel 3D image
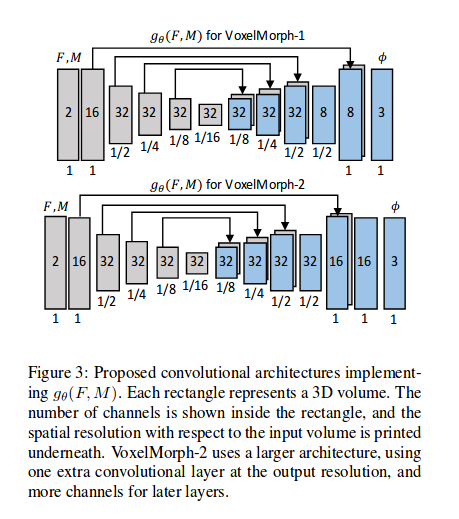
2 models
- VoxelMorph-1: One less layer at the final resolution and fewer channels in the last 3 layers
- VoxelMorph-2: Full network
The deformed image is computed using a differentiable operation based on spatial transformer networks (subpixel locations + interpolation)
Loss function
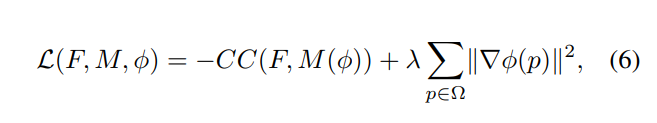
CC: Local cross-correlation (cross-correlation between images with local mean intensities subtracted out).
Regularization: L2 norm on the spatial gradients (approximated using differences between neighboring voxels). This enforces a spatially smooth deformation.
Experiments
Dataset: Multiple datasets are combined: ADNI, OASIS, ABIDE, ADHD200, MCIC, PPMI, HABS, Harvard GSP. All scans are resampled to 256x256x256, 1mm isotropic. FreeSurfer is used for affine spatial normalization and brain extraction, then images are cropped to 160x192x224. Dataset sizes (train/valid/test): 7329 / 250 / 250.
An atlas is used as the fixed image for all image pairs.
Test set ground truth is provided by expert-labeled anatomical segmentations.
Evaluation: Dice score: volume overlap of anatomical segmentations. Include any anatomical structures that are at least 100 voxels in volume for all test subjects, resulting in 29 structures.
Baseline: Symmetric Normalization (ANTs)
Implementation: Tensorflow using Keras.