FiberNET: An Ensemble Deep Learning Framework for Clustering White Matter Fibers
Summary
The authors propose a CNN architecture for white matter streamline classification (tractogram segmentation).
First, all subjects are registered to the standard MNI template so that they are all aligned with a shape-center. Then, they compute a spherical coordinate system in MNI space that is topologically equivalent to the brain volume.
All the streamlines are then converted to the spherical coordinate system.
(See the paper for the definition and computation of the potential equations).
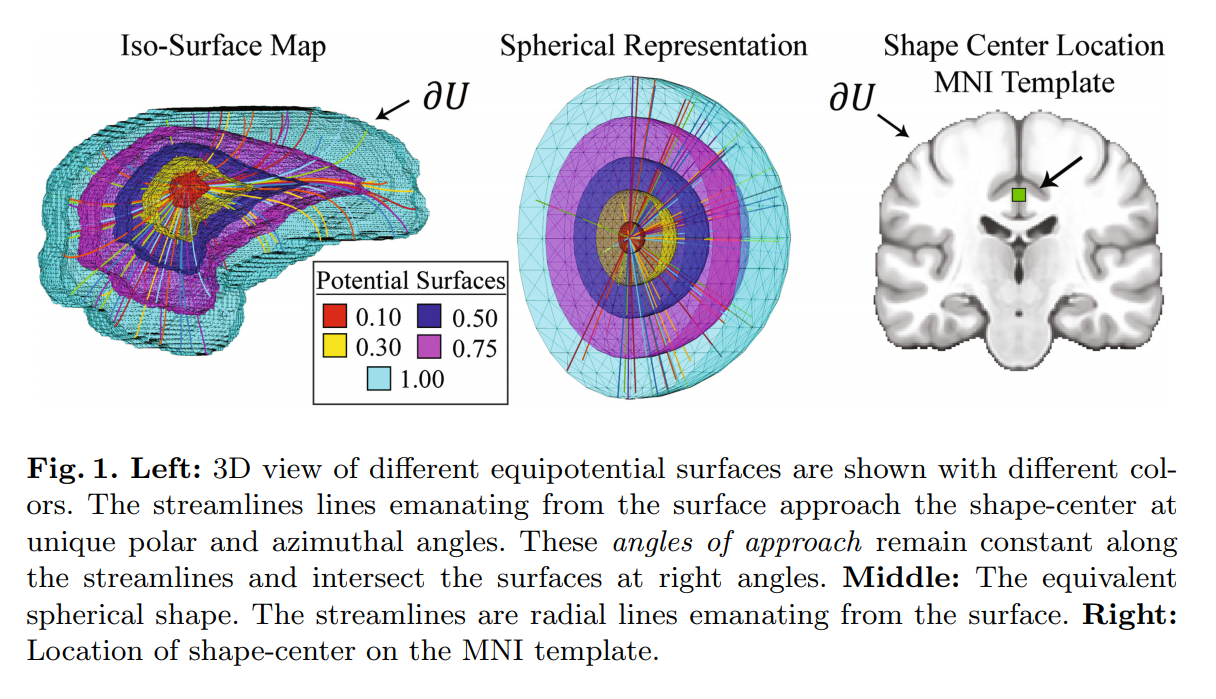
Streamlines
White matter bundles are usually uneven in the number of tracked streamlines. To address this problem, they 1) apply gaussian noise to the streamlines coordinates, and 2) reverse the order of points in the streamlines.
The authors tried resampling streamlines to 25, 50, 75 and 100 points, but noted that resampling over 50 points showed diminishing returns. Consequently, a streamline is represented as a 50 x 3 “image” or matrix.
Network
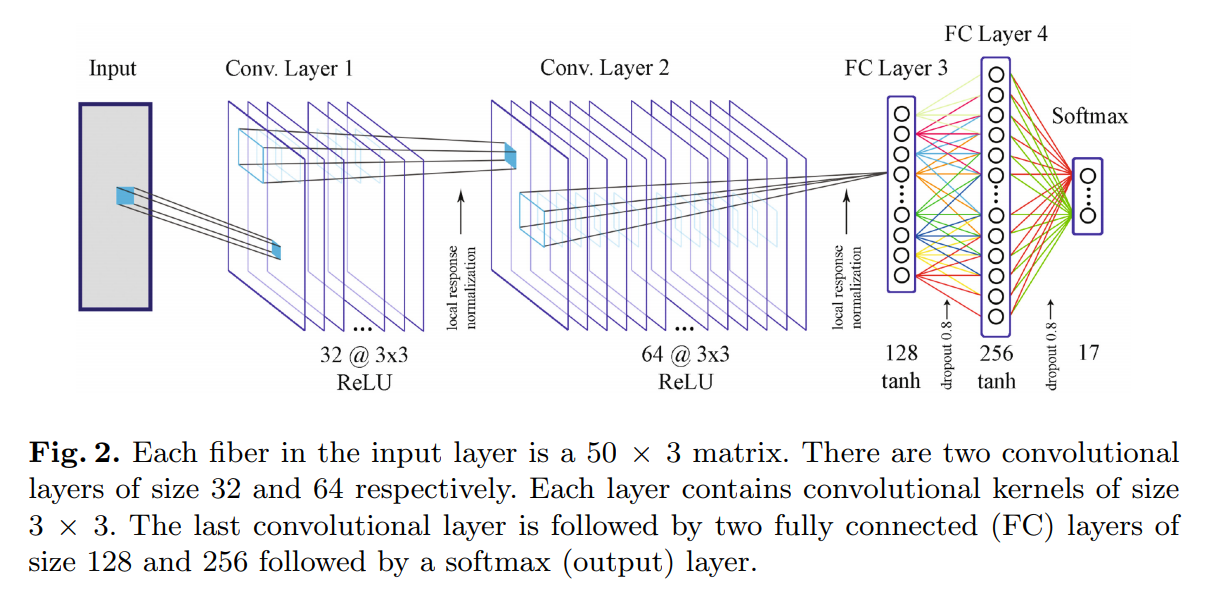
Streamlines must be classified as one of 17 anatomically relevant fiber bundles. Manual segmentation was done on 4 subjects to create a training set, and leave-one-out cross-validation was used during training. The authors also used bootstarpping to create 20 differents datasets, each with 600 000 streamlines. They trained 20 models to form an ensemble with majority voting at test time.
Results
The proposed method is compared to autoMATE, an existing automatic streamline clustering method (atlas-based method with ROI, length and distance criteria).
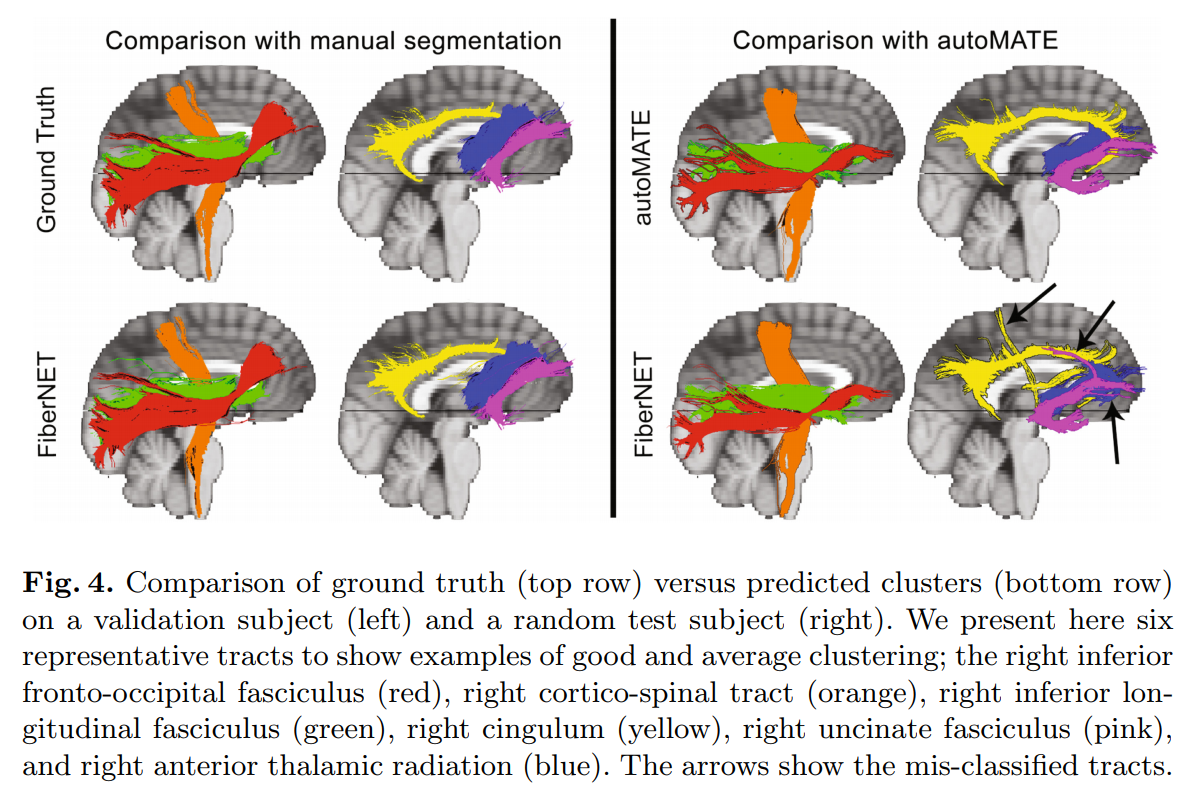
FiberNET doesn’t seem to perform as good as autoMATE, but the authors argue that using a CNN architecture is more flexible, and provides an opportunity to fix the misclassification problem (which isn’t possible with the atlas-based autoMATE).